
Novosphingobium sp. PhB165
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Novosphingobium; unclassified Novosphingobium
Average proteome isoelectric point is 6.35
Get precalculated fractions of proteins
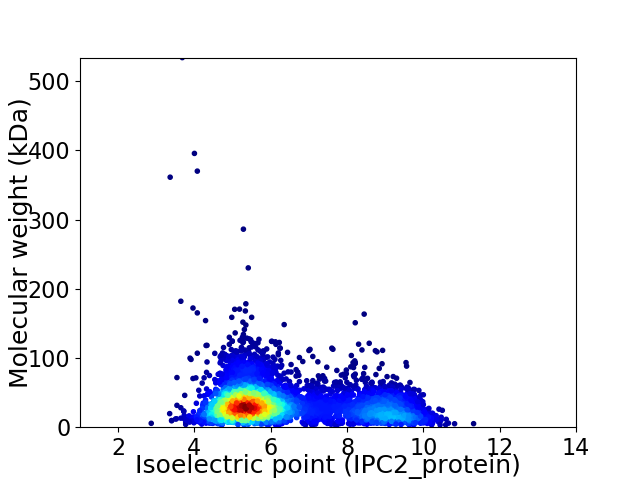
Virtual 2D-PAGE plot for 5096 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
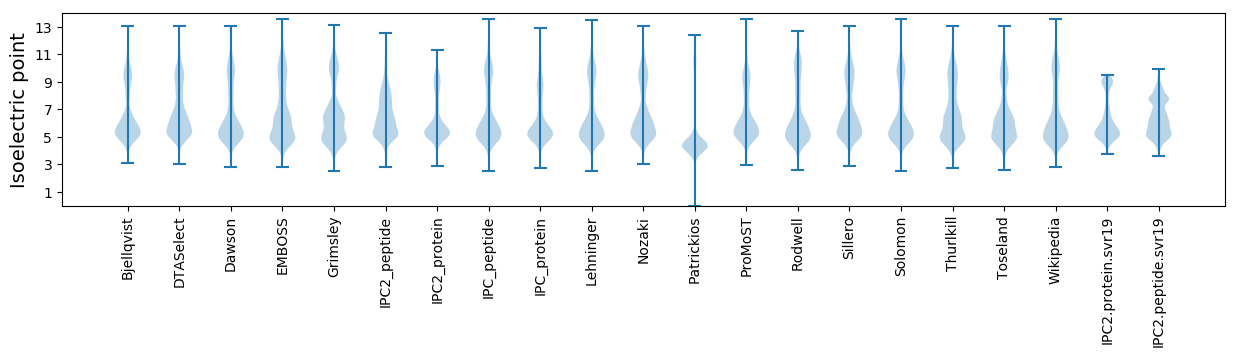
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4R1UXJ1|A0A4R1UXJ1_9SPHN Putative redox protein OS=Novosphingobium sp. PhB165 OX=2485105 GN=EDF56_101364 PE=4 SV=1
MM1 pKa = 7.01TARR4 pKa = 11.84LFALFATLAALLLAGTAQAQNATAAWSSLGLSNGGSVGSSTAITASDD51 pKa = 3.58GTTVSTAWSTTVGGAGTFAVASGYY75 pKa = 10.82NNYY78 pKa = 9.38VVYY81 pKa = 8.73NTPAIGGVSPGLLLNFDD98 pKa = 4.42ASRR101 pKa = 11.84FDD103 pKa = 4.05ANNKK107 pKa = 7.0ITMDD111 pKa = 2.9ITLNRR116 pKa = 11.84SVTGLAFSIADD127 pKa = 3.41IDD129 pKa = 5.16RR130 pKa = 11.84EE131 pKa = 4.65TTGNNYY137 pKa = 9.48NRR139 pKa = 11.84DD140 pKa = 3.77AVAVYY145 pKa = 10.56YY146 pKa = 8.61DD147 pKa = 3.62TGNGTFVNAVSNSAFWSDD165 pKa = 3.41GSAVTNDD172 pKa = 3.04TSGWNGTAASANNSTNGTLTFNFGTTAVKK201 pKa = 10.35RR202 pKa = 11.84IRR204 pKa = 11.84IVYY207 pKa = 9.84NSYY210 pKa = 8.74TVSGTANPTAQFIVLSGLAFNAKK233 pKa = 9.99GADD236 pKa = 3.44LSLGNTLLTSSPTSGGLATFRR257 pKa = 11.84LTVTNASTSTDD268 pKa = 2.92TATDD272 pKa = 3.45VKK274 pKa = 10.64VTDD277 pKa = 3.83TLPDD281 pKa = 3.14GFTYY285 pKa = 10.62VSSNGTGSFNAATGVWTVGSIAPGASVTLDD315 pKa = 2.86ITGTVNASAGATLTNTAEE333 pKa = 4.23ITASDD338 pKa = 3.86QNDD341 pKa = 3.82PDD343 pKa = 3.98STPGNGVTSEE353 pKa = 4.66DD354 pKa = 4.48DD355 pKa = 3.63YY356 pKa = 12.26ASATLTVTGSRR367 pKa = 11.84VAGTPPTLSCAAGTVVFDD385 pKa = 3.63WSNRR389 pKa = 11.84AWTAGSTSNSYY400 pKa = 11.31SFDD403 pKa = 3.47TLGTISFALANQGTWLSSSTYY424 pKa = 10.15GGQSPVRR431 pKa = 11.84QNVVNGGLNDD441 pKa = 3.5YY442 pKa = 11.49ALFEE446 pKa = 4.44MVNMPNISAEE456 pKa = 4.04ATTTITLPTALPGAQFKK473 pKa = 10.87IFDD476 pKa = 3.34VDD478 pKa = 3.9YY479 pKa = 11.34YY480 pKa = 11.29SGQFADD486 pKa = 3.57KK487 pKa = 9.14VTVEE491 pKa = 3.79GRR493 pKa = 11.84YY494 pKa = 9.74NGAVVTPTLTNGVSNYY510 pKa = 10.32VIGNSAYY517 pKa = 10.51GDD519 pKa = 4.04GLSDD523 pKa = 4.49DD524 pKa = 4.55ASADD528 pKa = 3.39GTVTVTFSQPIDD540 pKa = 3.78TIIVRR545 pKa = 11.84YY546 pKa = 9.09GNHH549 pKa = 5.69SMAPSDD555 pKa = 4.23PGQQAIALHH564 pKa = 7.4DD565 pKa = 3.4ITFCRR570 pKa = 11.84PSTTLAVSKK579 pKa = 9.53TSSVVSDD586 pKa = 3.89PVSGTNNPKK595 pKa = 10.34AIPGATVRR603 pKa = 11.84YY604 pKa = 8.83CILVANTGSSAATSVAASDD623 pKa = 5.24PIPATLTYY631 pKa = 10.2VAGSIYY637 pKa = 10.53SGSDD641 pKa = 3.11CASANTAEE649 pKa = 5.09DD650 pKa = 4.52DD651 pKa = 3.7NDD653 pKa = 4.12TGSDD657 pKa = 3.32EE658 pKa = 4.29SDD660 pKa = 3.48PYY662 pKa = 9.82GASYY666 pKa = 11.5ASGTVKK672 pKa = 10.67ASAPTLAAGTSFAIRR687 pKa = 11.84FDD689 pKa = 3.42ATVNN693 pKa = 3.43
MM1 pKa = 7.01TARR4 pKa = 11.84LFALFATLAALLLAGTAQAQNATAAWSSLGLSNGGSVGSSTAITASDD51 pKa = 3.58GTTVSTAWSTTVGGAGTFAVASGYY75 pKa = 10.82NNYY78 pKa = 9.38VVYY81 pKa = 8.73NTPAIGGVSPGLLLNFDD98 pKa = 4.42ASRR101 pKa = 11.84FDD103 pKa = 4.05ANNKK107 pKa = 7.0ITMDD111 pKa = 2.9ITLNRR116 pKa = 11.84SVTGLAFSIADD127 pKa = 3.41IDD129 pKa = 5.16RR130 pKa = 11.84EE131 pKa = 4.65TTGNNYY137 pKa = 9.48NRR139 pKa = 11.84DD140 pKa = 3.77AVAVYY145 pKa = 10.56YY146 pKa = 8.61DD147 pKa = 3.62TGNGTFVNAVSNSAFWSDD165 pKa = 3.41GSAVTNDD172 pKa = 3.04TSGWNGTAASANNSTNGTLTFNFGTTAVKK201 pKa = 10.35RR202 pKa = 11.84IRR204 pKa = 11.84IVYY207 pKa = 9.84NSYY210 pKa = 8.74TVSGTANPTAQFIVLSGLAFNAKK233 pKa = 9.99GADD236 pKa = 3.44LSLGNTLLTSSPTSGGLATFRR257 pKa = 11.84LTVTNASTSTDD268 pKa = 2.92TATDD272 pKa = 3.45VKK274 pKa = 10.64VTDD277 pKa = 3.83TLPDD281 pKa = 3.14GFTYY285 pKa = 10.62VSSNGTGSFNAATGVWTVGSIAPGASVTLDD315 pKa = 2.86ITGTVNASAGATLTNTAEE333 pKa = 4.23ITASDD338 pKa = 3.86QNDD341 pKa = 3.82PDD343 pKa = 3.98STPGNGVTSEE353 pKa = 4.66DD354 pKa = 4.48DD355 pKa = 3.63YY356 pKa = 12.26ASATLTVTGSRR367 pKa = 11.84VAGTPPTLSCAAGTVVFDD385 pKa = 3.63WSNRR389 pKa = 11.84AWTAGSTSNSYY400 pKa = 11.31SFDD403 pKa = 3.47TLGTISFALANQGTWLSSSTYY424 pKa = 10.15GGQSPVRR431 pKa = 11.84QNVVNGGLNDD441 pKa = 3.5YY442 pKa = 11.49ALFEE446 pKa = 4.44MVNMPNISAEE456 pKa = 4.04ATTTITLPTALPGAQFKK473 pKa = 10.87IFDD476 pKa = 3.34VDD478 pKa = 3.9YY479 pKa = 11.34YY480 pKa = 11.29SGQFADD486 pKa = 3.57KK487 pKa = 9.14VTVEE491 pKa = 3.79GRR493 pKa = 11.84YY494 pKa = 9.74NGAVVTPTLTNGVSNYY510 pKa = 10.32VIGNSAYY517 pKa = 10.51GDD519 pKa = 4.04GLSDD523 pKa = 4.49DD524 pKa = 4.55ASADD528 pKa = 3.39GTVTVTFSQPIDD540 pKa = 3.78TIIVRR545 pKa = 11.84YY546 pKa = 9.09GNHH549 pKa = 5.69SMAPSDD555 pKa = 4.23PGQQAIALHH564 pKa = 7.4DD565 pKa = 3.4ITFCRR570 pKa = 11.84PSTTLAVSKK579 pKa = 9.53TSSVVSDD586 pKa = 3.89PVSGTNNPKK595 pKa = 10.34AIPGATVRR603 pKa = 11.84YY604 pKa = 8.83CILVANTGSSAATSVAASDD623 pKa = 5.24PIPATLTYY631 pKa = 10.2VAGSIYY637 pKa = 10.53SGSDD641 pKa = 3.11CASANTAEE649 pKa = 5.09DD650 pKa = 4.52DD651 pKa = 3.7NDD653 pKa = 4.12TGSDD657 pKa = 3.32EE658 pKa = 4.29SDD660 pKa = 3.48PYY662 pKa = 9.82GASYY666 pKa = 11.5ASGTVKK672 pKa = 10.67ASAPTLAAGTSFAIRR687 pKa = 11.84FDD689 pKa = 3.42ATVNN693 pKa = 3.43
Molecular weight: 70.37 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4R1UQE9|A0A4R1UQE9_9SPHN Two-component system chemotaxis response regulator CheY OS=Novosphingobium sp. PhB165 OX=2485105 GN=EDF56_103211 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.54GFRR19 pKa = 11.84ARR21 pKa = 11.84TATVGGRR28 pKa = 11.84KK29 pKa = 8.17VLRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.54GFRR19 pKa = 11.84ARR21 pKa = 11.84TATVGGRR28 pKa = 11.84KK29 pKa = 8.17VLRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
Molecular weight: 5.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1697015 |
29 |
6018 |
333.0 |
35.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.998 ± 0.052 | 0.823 ± 0.011 |
5.822 ± 0.026 | 5.447 ± 0.041 |
3.566 ± 0.021 | 9.074 ± 0.084 |
2.072 ± 0.017 | 4.759 ± 0.022 |
2.941 ± 0.026 | 9.779 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.432 ± 0.021 | 2.684 ± 0.027 |
5.375 ± 0.028 | 3.173 ± 0.019 |
6.948 ± 0.051 | 5.589 ± 0.034 |
5.49 ± 0.05 | 7.271 ± 0.027 |
1.476 ± 0.015 | 2.282 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
