
Capybara microvirus Cap1_SP_52
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.12
Get precalculated fractions of proteins
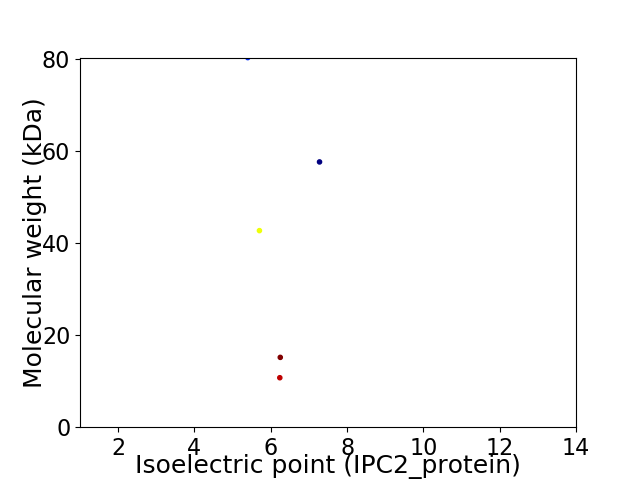
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4V1FVM6|A0A4V1FVM6_9VIRU Major capsid protein OS=Capybara microvirus Cap1_SP_52 OX=2584782 PE=3 SV=1
MM1 pKa = 7.6SKK3 pKa = 10.62NPFNIKK9 pKa = 9.79DD10 pKa = 3.61VRR12 pKa = 11.84NHH14 pKa = 5.54VSHH17 pKa = 6.96NGFDD21 pKa = 3.72LSEE24 pKa = 4.43KK25 pKa = 10.7KK26 pKa = 10.77LFTAKK31 pKa = 10.32CGEE34 pKa = 4.76LLPIMHH40 pKa = 6.52RR41 pKa = 11.84TVMPGDD47 pKa = 3.51KK48 pKa = 10.42FRR50 pKa = 11.84ITKK53 pKa = 9.15QHH55 pKa = 5.58LTRR58 pKa = 11.84TIPVDD63 pKa = 3.12TAAYY67 pKa = 6.79TRR69 pKa = 11.84LRR71 pKa = 11.84EE72 pKa = 3.86HH73 pKa = 7.01FEE75 pKa = 4.27FFFVPYY81 pKa = 10.04DD82 pKa = 3.47QLYY85 pKa = 9.76RR86 pKa = 11.84PANQVLEE93 pKa = 4.22NLSGLSSTRR102 pKa = 11.84ATSLVEE108 pKa = 3.68YY109 pKa = 8.83TQVNEE114 pKa = 3.86YY115 pKa = 10.19LPNIVYY121 pKa = 9.86SAPQSQEE128 pKa = 3.26TSYY131 pKa = 11.27LALFPSGTQSFDD143 pKa = 2.9NTTPKK148 pKa = 10.69PEE150 pKa = 4.0FRR152 pKa = 11.84NDD154 pKa = 4.37LITSLASMLNNVDD167 pKa = 2.82VSSRR171 pKa = 11.84YY172 pKa = 10.03NEE174 pKa = 3.91VGRR177 pKa = 11.84NPIYY181 pKa = 10.34ATIKK185 pKa = 9.41LLNYY189 pKa = 10.16LGYY192 pKa = 10.33GYY194 pKa = 10.46HH195 pKa = 7.22ADD197 pKa = 3.42SFLNEE202 pKa = 3.69IQPYY206 pKa = 8.86ISKK209 pKa = 10.68DD210 pKa = 3.14GEE212 pKa = 4.43RR213 pKa = 11.84ILGKK217 pKa = 10.53SFDD220 pKa = 3.78FGVAKK225 pKa = 9.19TQTRR229 pKa = 11.84LSLLPLAAYY238 pKa = 9.28QKK240 pKa = 10.47IYY242 pKa = 10.41FDD244 pKa = 4.05YY245 pKa = 10.76YY246 pKa = 11.14RR247 pKa = 11.84NEE249 pKa = 3.7QWEE252 pKa = 4.34KK253 pKa = 11.49DD254 pKa = 3.66NPSCYY259 pKa = 10.36NFDD262 pKa = 3.32YY263 pKa = 10.58MMPHH267 pKa = 6.74INVDD271 pKa = 3.22SSLNNVMQTTTTQCTLLKK289 pKa = 8.76THH291 pKa = 7.2ASFGSSPSVSFIDD304 pKa = 3.9FAFNNFATLRR314 pKa = 11.84YY315 pKa = 9.84RR316 pKa = 11.84NFKK319 pKa = 10.05KK320 pKa = 10.65DD321 pKa = 3.6LFFGCLPRR329 pKa = 11.84PQAGNTTFVPTSPIFKK345 pKa = 9.36ATDD348 pKa = 3.24TEE350 pKa = 4.67FPEE353 pKa = 3.91NGYY356 pKa = 9.94NRR358 pKa = 11.84DD359 pKa = 3.06VNTTYY364 pKa = 10.45FRR366 pKa = 11.84NANALFQSFPRR377 pKa = 11.84LQNLTPEE384 pKa = 4.1QQNIEE389 pKa = 4.15GYY391 pKa = 10.7LYY393 pKa = 10.9DD394 pKa = 3.67GSNYY398 pKa = 7.28VTPATGGFAIQDD410 pKa = 3.51FRR412 pKa = 11.84IGMALQRR419 pKa = 11.84YY420 pKa = 8.32RR421 pKa = 11.84EE422 pKa = 4.21VVGSARR428 pKa = 11.84HH429 pKa = 6.01DD430 pKa = 3.52FVNEE434 pKa = 3.74TEE436 pKa = 4.08RR437 pKa = 11.84QFGVKK442 pKa = 9.55VPQYY446 pKa = 10.44LSHH449 pKa = 6.97KK450 pKa = 9.24VQFLGEE456 pKa = 3.9FSNWIGISAIVNNNLDD472 pKa = 4.03TADD475 pKa = 3.6STTDD479 pKa = 2.81IKK481 pKa = 11.38GIGVGSSSDD490 pKa = 3.05NDD492 pKa = 4.27AINFKK497 pKa = 11.0AMEE500 pKa = 4.58FGIIMCIYY508 pKa = 10.03SVEE511 pKa = 4.07PQVDD515 pKa = 3.72YY516 pKa = 11.32EE517 pKa = 4.5FDD519 pKa = 3.51GFAPDD524 pKa = 4.49LLKK527 pKa = 11.1CHH529 pKa = 6.76ASSFANPAFDD539 pKa = 4.22NIGYY543 pKa = 9.92EE544 pKa = 4.47PIPKK548 pKa = 10.12ALISDD553 pKa = 3.78EE554 pKa = 4.4QYY556 pKa = 11.56GSDD559 pKa = 3.03IDD561 pKa = 4.18MPNQYY566 pKa = 10.2HH567 pKa = 5.49SQNHH571 pKa = 5.91LGFNVRR577 pKa = 11.84YY578 pKa = 8.53WDD580 pKa = 3.7YY581 pKa = 10.73KK582 pKa = 9.64TGFDD586 pKa = 3.55NVLGAFRR593 pKa = 11.84EE594 pKa = 4.42TLVTWVAPLNFNTLLSQLVPNFGDD618 pKa = 3.64ASDD621 pKa = 4.66ANTARR626 pKa = 11.84VNSHH630 pKa = 5.76ISANGNMKK638 pKa = 9.93YY639 pKa = 10.37HH640 pKa = 6.54NNPNYY645 pKa = 10.37GYY647 pKa = 9.99SQFPGYY653 pKa = 10.35GYY655 pKa = 9.6PIINYY660 pKa = 9.57AVFKK664 pKa = 10.55INPSLLDD671 pKa = 3.64TIFFAQVNDD680 pKa = 3.8LTEE683 pKa = 4.1TDD685 pKa = 3.56QFLVNCNLSIKK696 pKa = 10.3ALRR699 pKa = 11.84PLSWSGLPYY708 pKa = 10.89
MM1 pKa = 7.6SKK3 pKa = 10.62NPFNIKK9 pKa = 9.79DD10 pKa = 3.61VRR12 pKa = 11.84NHH14 pKa = 5.54VSHH17 pKa = 6.96NGFDD21 pKa = 3.72LSEE24 pKa = 4.43KK25 pKa = 10.7KK26 pKa = 10.77LFTAKK31 pKa = 10.32CGEE34 pKa = 4.76LLPIMHH40 pKa = 6.52RR41 pKa = 11.84TVMPGDD47 pKa = 3.51KK48 pKa = 10.42FRR50 pKa = 11.84ITKK53 pKa = 9.15QHH55 pKa = 5.58LTRR58 pKa = 11.84TIPVDD63 pKa = 3.12TAAYY67 pKa = 6.79TRR69 pKa = 11.84LRR71 pKa = 11.84EE72 pKa = 3.86HH73 pKa = 7.01FEE75 pKa = 4.27FFFVPYY81 pKa = 10.04DD82 pKa = 3.47QLYY85 pKa = 9.76RR86 pKa = 11.84PANQVLEE93 pKa = 4.22NLSGLSSTRR102 pKa = 11.84ATSLVEE108 pKa = 3.68YY109 pKa = 8.83TQVNEE114 pKa = 3.86YY115 pKa = 10.19LPNIVYY121 pKa = 9.86SAPQSQEE128 pKa = 3.26TSYY131 pKa = 11.27LALFPSGTQSFDD143 pKa = 2.9NTTPKK148 pKa = 10.69PEE150 pKa = 4.0FRR152 pKa = 11.84NDD154 pKa = 4.37LITSLASMLNNVDD167 pKa = 2.82VSSRR171 pKa = 11.84YY172 pKa = 10.03NEE174 pKa = 3.91VGRR177 pKa = 11.84NPIYY181 pKa = 10.34ATIKK185 pKa = 9.41LLNYY189 pKa = 10.16LGYY192 pKa = 10.33GYY194 pKa = 10.46HH195 pKa = 7.22ADD197 pKa = 3.42SFLNEE202 pKa = 3.69IQPYY206 pKa = 8.86ISKK209 pKa = 10.68DD210 pKa = 3.14GEE212 pKa = 4.43RR213 pKa = 11.84ILGKK217 pKa = 10.53SFDD220 pKa = 3.78FGVAKK225 pKa = 9.19TQTRR229 pKa = 11.84LSLLPLAAYY238 pKa = 9.28QKK240 pKa = 10.47IYY242 pKa = 10.41FDD244 pKa = 4.05YY245 pKa = 10.76YY246 pKa = 11.14RR247 pKa = 11.84NEE249 pKa = 3.7QWEE252 pKa = 4.34KK253 pKa = 11.49DD254 pKa = 3.66NPSCYY259 pKa = 10.36NFDD262 pKa = 3.32YY263 pKa = 10.58MMPHH267 pKa = 6.74INVDD271 pKa = 3.22SSLNNVMQTTTTQCTLLKK289 pKa = 8.76THH291 pKa = 7.2ASFGSSPSVSFIDD304 pKa = 3.9FAFNNFATLRR314 pKa = 11.84YY315 pKa = 9.84RR316 pKa = 11.84NFKK319 pKa = 10.05KK320 pKa = 10.65DD321 pKa = 3.6LFFGCLPRR329 pKa = 11.84PQAGNTTFVPTSPIFKK345 pKa = 9.36ATDD348 pKa = 3.24TEE350 pKa = 4.67FPEE353 pKa = 3.91NGYY356 pKa = 9.94NRR358 pKa = 11.84DD359 pKa = 3.06VNTTYY364 pKa = 10.45FRR366 pKa = 11.84NANALFQSFPRR377 pKa = 11.84LQNLTPEE384 pKa = 4.1QQNIEE389 pKa = 4.15GYY391 pKa = 10.7LYY393 pKa = 10.9DD394 pKa = 3.67GSNYY398 pKa = 7.28VTPATGGFAIQDD410 pKa = 3.51FRR412 pKa = 11.84IGMALQRR419 pKa = 11.84YY420 pKa = 8.32RR421 pKa = 11.84EE422 pKa = 4.21VVGSARR428 pKa = 11.84HH429 pKa = 6.01DD430 pKa = 3.52FVNEE434 pKa = 3.74TEE436 pKa = 4.08RR437 pKa = 11.84QFGVKK442 pKa = 9.55VPQYY446 pKa = 10.44LSHH449 pKa = 6.97KK450 pKa = 9.24VQFLGEE456 pKa = 3.9FSNWIGISAIVNNNLDD472 pKa = 4.03TADD475 pKa = 3.6STTDD479 pKa = 2.81IKK481 pKa = 11.38GIGVGSSSDD490 pKa = 3.05NDD492 pKa = 4.27AINFKK497 pKa = 11.0AMEE500 pKa = 4.58FGIIMCIYY508 pKa = 10.03SVEE511 pKa = 4.07PQVDD515 pKa = 3.72YY516 pKa = 11.32EE517 pKa = 4.5FDD519 pKa = 3.51GFAPDD524 pKa = 4.49LLKK527 pKa = 11.1CHH529 pKa = 6.76ASSFANPAFDD539 pKa = 4.22NIGYY543 pKa = 9.92EE544 pKa = 4.47PIPKK548 pKa = 10.12ALISDD553 pKa = 3.78EE554 pKa = 4.4QYY556 pKa = 11.56GSDD559 pKa = 3.03IDD561 pKa = 4.18MPNQYY566 pKa = 10.2HH567 pKa = 5.49SQNHH571 pKa = 5.91LGFNVRR577 pKa = 11.84YY578 pKa = 8.53WDD580 pKa = 3.7YY581 pKa = 10.73KK582 pKa = 9.64TGFDD586 pKa = 3.55NVLGAFRR593 pKa = 11.84EE594 pKa = 4.42TLVTWVAPLNFNTLLSQLVPNFGDD618 pKa = 3.64ASDD621 pKa = 4.66ANTARR626 pKa = 11.84VNSHH630 pKa = 5.76ISANGNMKK638 pKa = 9.93YY639 pKa = 10.37HH640 pKa = 6.54NNPNYY645 pKa = 10.37GYY647 pKa = 9.99SQFPGYY653 pKa = 10.35GYY655 pKa = 9.6PIINYY660 pKa = 9.57AVFKK664 pKa = 10.55INPSLLDD671 pKa = 3.64TIFFAQVNDD680 pKa = 3.8LTEE683 pKa = 4.1TDD685 pKa = 3.56QFLVNCNLSIKK696 pKa = 10.3ALRR699 pKa = 11.84PLSWSGLPYY708 pKa = 10.89
Molecular weight: 80.4 kDa
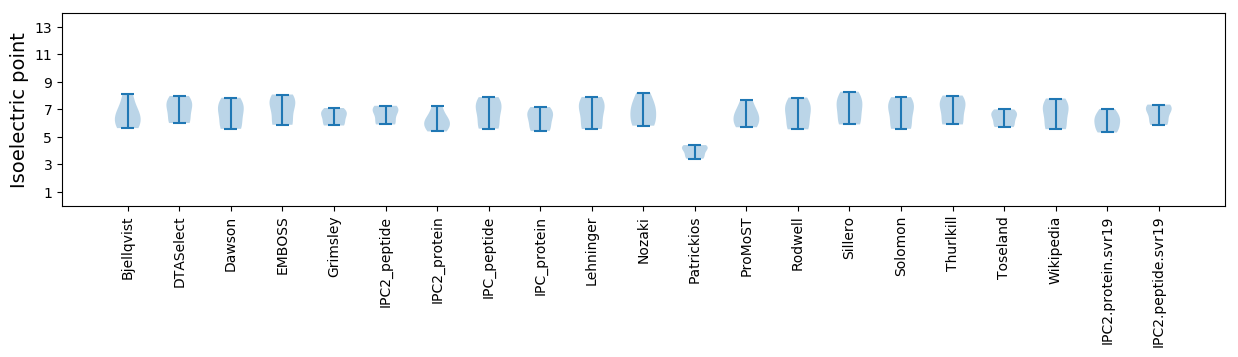
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W3R5|A0A4P8W3R5_9VIRU Uncharacterized protein OS=Capybara microvirus Cap1_SP_52 OX=2584782 PE=4 SV=1
MM1 pKa = 7.36SLCLKK6 pKa = 9.77PIKK9 pKa = 9.74IWQNNRR15 pKa = 11.84EE16 pKa = 4.07RR17 pKa = 11.84TVSCGHH23 pKa = 6.88CAHH26 pKa = 7.49CINAKK31 pKa = 10.03HH32 pKa = 6.11IRR34 pKa = 11.84NKK36 pKa = 10.24QLVEE40 pKa = 4.17LEE42 pKa = 4.85SNSHH46 pKa = 6.51PYY48 pKa = 10.68EE49 pKa = 4.17LFCTLTYY56 pKa = 11.0DD57 pKa = 4.07DD58 pKa = 3.8YY59 pKa = 11.5HH60 pKa = 7.92VPKK63 pKa = 10.47VHH65 pKa = 6.62YY66 pKa = 10.43SHH68 pKa = 6.99TNKK71 pKa = 10.2LSSIEE76 pKa = 4.14SEE78 pKa = 5.05DD79 pKa = 2.97IFEE82 pKa = 4.35FTNSRR87 pKa = 11.84FKK89 pKa = 11.49DD90 pKa = 4.09EE91 pKa = 4.14VCKK94 pKa = 10.92GRR96 pKa = 11.84FAVPNYY102 pKa = 9.04YY103 pKa = 10.66LKK105 pKa = 10.68DD106 pKa = 3.39VHH108 pKa = 7.11SINMRR113 pKa = 11.84QQLFNGEE120 pKa = 4.08IRR122 pKa = 11.84TLNYY126 pKa = 10.08VDD128 pKa = 4.48LRR130 pKa = 11.84LYY132 pKa = 9.55LQRR135 pKa = 11.84VRR137 pKa = 11.84QALRR141 pKa = 11.84RR142 pKa = 11.84SLGTNVRR149 pKa = 11.84FYY151 pKa = 11.6AVGEE155 pKa = 4.08YY156 pKa = 10.64GPKK159 pKa = 9.89KK160 pKa = 10.25YY161 pKa = 9.92RR162 pKa = 11.84PHH164 pKa = 6.62YY165 pKa = 10.21HH166 pKa = 6.68IIFWCDD172 pKa = 3.03DD173 pKa = 3.56LASRR177 pKa = 11.84DD178 pKa = 3.66WLLVNALSYY187 pKa = 9.69WPYY190 pKa = 8.59GTQIEE195 pKa = 4.49IEE197 pKa = 4.14QSRR200 pKa = 11.84KK201 pKa = 10.19KK202 pKa = 10.47LDD204 pKa = 3.44SYY206 pKa = 11.2LAGYY210 pKa = 9.46VNSEE214 pKa = 3.93TMLPPFFSCLKK225 pKa = 10.53SPMRR229 pKa = 11.84PKK231 pKa = 10.4AYY233 pKa = 9.82HH234 pKa = 6.48SNFLGQTYY242 pKa = 9.34PFAYY246 pKa = 9.64LSVMFHH252 pKa = 6.89SDD254 pKa = 3.02PGKK257 pKa = 10.48KK258 pKa = 9.64FEE260 pKa = 5.25LFKK263 pKa = 8.64TTKK266 pKa = 10.33GIYY269 pKa = 8.8DD270 pKa = 4.89TIPKK274 pKa = 10.33AIFDD278 pKa = 3.66YY279 pKa = 10.19FYY281 pKa = 11.16RR282 pKa = 11.84KK283 pKa = 9.69LPNYY287 pKa = 9.88QNLDD291 pKa = 3.34NHH293 pKa = 6.52EE294 pKa = 4.17KK295 pKa = 7.1TTLYY299 pKa = 11.27KK300 pKa = 10.55LFNVMYY306 pKa = 9.8DD307 pKa = 3.48YY308 pKa = 10.79RR309 pKa = 11.84QKK311 pKa = 10.79QVGEE315 pKa = 4.27NIPLEE320 pKa = 4.18NYY322 pKa = 9.6KK323 pKa = 10.51FSAFLEE329 pKa = 4.16DD330 pKa = 4.62LKK332 pKa = 11.35DD333 pKa = 3.86FEE335 pKa = 5.54RR336 pKa = 11.84NEE338 pKa = 4.01FYY340 pKa = 10.35IINLLGIDD348 pKa = 5.1DD349 pKa = 4.09FFEE352 pKa = 4.1YY353 pKa = 10.65LKK355 pKa = 10.41PLEE358 pKa = 4.03VNYY361 pKa = 10.2EE362 pKa = 4.15YY363 pKa = 11.35KK364 pKa = 10.67CVGEE368 pKa = 4.11NLLTYY373 pKa = 9.91RR374 pKa = 11.84WHH376 pKa = 7.08PKK378 pKa = 10.03LISLYY383 pKa = 10.01RR384 pKa = 11.84VFSIGKK390 pKa = 8.78FYY392 pKa = 10.73FWLSDD397 pKa = 3.18ITSINYY403 pKa = 7.41NTLIDD408 pKa = 3.66KK409 pKa = 10.11VIYY412 pKa = 10.68YY413 pKa = 9.89FDD415 pKa = 4.27KK416 pKa = 10.87EE417 pKa = 4.31QPQQMLANFFSNIEE431 pKa = 4.14TLPDD435 pKa = 3.77ISPQDD440 pKa = 3.48LSFLYY445 pKa = 10.93NLDD448 pKa = 4.71DD449 pKa = 5.05DD450 pKa = 5.12YY451 pKa = 11.89KK452 pKa = 11.19DD453 pKa = 3.3SKK455 pKa = 11.06LYY457 pKa = 10.69QCIKK461 pKa = 7.87THH463 pKa = 6.71AIEE466 pKa = 4.19VANDD470 pKa = 3.33NVKK473 pKa = 10.28HH474 pKa = 6.11KK475 pKa = 10.87KK476 pKa = 10.18EE477 pKa = 3.86NDD479 pKa = 3.38GLKK482 pKa = 9.93II483 pKa = 3.77
MM1 pKa = 7.36SLCLKK6 pKa = 9.77PIKK9 pKa = 9.74IWQNNRR15 pKa = 11.84EE16 pKa = 4.07RR17 pKa = 11.84TVSCGHH23 pKa = 6.88CAHH26 pKa = 7.49CINAKK31 pKa = 10.03HH32 pKa = 6.11IRR34 pKa = 11.84NKK36 pKa = 10.24QLVEE40 pKa = 4.17LEE42 pKa = 4.85SNSHH46 pKa = 6.51PYY48 pKa = 10.68EE49 pKa = 4.17LFCTLTYY56 pKa = 11.0DD57 pKa = 4.07DD58 pKa = 3.8YY59 pKa = 11.5HH60 pKa = 7.92VPKK63 pKa = 10.47VHH65 pKa = 6.62YY66 pKa = 10.43SHH68 pKa = 6.99TNKK71 pKa = 10.2LSSIEE76 pKa = 4.14SEE78 pKa = 5.05DD79 pKa = 2.97IFEE82 pKa = 4.35FTNSRR87 pKa = 11.84FKK89 pKa = 11.49DD90 pKa = 4.09EE91 pKa = 4.14VCKK94 pKa = 10.92GRR96 pKa = 11.84FAVPNYY102 pKa = 9.04YY103 pKa = 10.66LKK105 pKa = 10.68DD106 pKa = 3.39VHH108 pKa = 7.11SINMRR113 pKa = 11.84QQLFNGEE120 pKa = 4.08IRR122 pKa = 11.84TLNYY126 pKa = 10.08VDD128 pKa = 4.48LRR130 pKa = 11.84LYY132 pKa = 9.55LQRR135 pKa = 11.84VRR137 pKa = 11.84QALRR141 pKa = 11.84RR142 pKa = 11.84SLGTNVRR149 pKa = 11.84FYY151 pKa = 11.6AVGEE155 pKa = 4.08YY156 pKa = 10.64GPKK159 pKa = 9.89KK160 pKa = 10.25YY161 pKa = 9.92RR162 pKa = 11.84PHH164 pKa = 6.62YY165 pKa = 10.21HH166 pKa = 6.68IIFWCDD172 pKa = 3.03DD173 pKa = 3.56LASRR177 pKa = 11.84DD178 pKa = 3.66WLLVNALSYY187 pKa = 9.69WPYY190 pKa = 8.59GTQIEE195 pKa = 4.49IEE197 pKa = 4.14QSRR200 pKa = 11.84KK201 pKa = 10.19KK202 pKa = 10.47LDD204 pKa = 3.44SYY206 pKa = 11.2LAGYY210 pKa = 9.46VNSEE214 pKa = 3.93TMLPPFFSCLKK225 pKa = 10.53SPMRR229 pKa = 11.84PKK231 pKa = 10.4AYY233 pKa = 9.82HH234 pKa = 6.48SNFLGQTYY242 pKa = 9.34PFAYY246 pKa = 9.64LSVMFHH252 pKa = 6.89SDD254 pKa = 3.02PGKK257 pKa = 10.48KK258 pKa = 9.64FEE260 pKa = 5.25LFKK263 pKa = 8.64TTKK266 pKa = 10.33GIYY269 pKa = 8.8DD270 pKa = 4.89TIPKK274 pKa = 10.33AIFDD278 pKa = 3.66YY279 pKa = 10.19FYY281 pKa = 11.16RR282 pKa = 11.84KK283 pKa = 9.69LPNYY287 pKa = 9.88QNLDD291 pKa = 3.34NHH293 pKa = 6.52EE294 pKa = 4.17KK295 pKa = 7.1TTLYY299 pKa = 11.27KK300 pKa = 10.55LFNVMYY306 pKa = 9.8DD307 pKa = 3.48YY308 pKa = 10.79RR309 pKa = 11.84QKK311 pKa = 10.79QVGEE315 pKa = 4.27NIPLEE320 pKa = 4.18NYY322 pKa = 9.6KK323 pKa = 10.51FSAFLEE329 pKa = 4.16DD330 pKa = 4.62LKK332 pKa = 11.35DD333 pKa = 3.86FEE335 pKa = 5.54RR336 pKa = 11.84NEE338 pKa = 4.01FYY340 pKa = 10.35IINLLGIDD348 pKa = 5.1DD349 pKa = 4.09FFEE352 pKa = 4.1YY353 pKa = 10.65LKK355 pKa = 10.41PLEE358 pKa = 4.03VNYY361 pKa = 10.2EE362 pKa = 4.15YY363 pKa = 11.35KK364 pKa = 10.67CVGEE368 pKa = 4.11NLLTYY373 pKa = 9.91RR374 pKa = 11.84WHH376 pKa = 7.08PKK378 pKa = 10.03LISLYY383 pKa = 10.01RR384 pKa = 11.84VFSIGKK390 pKa = 8.78FYY392 pKa = 10.73FWLSDD397 pKa = 3.18ITSINYY403 pKa = 7.41NTLIDD408 pKa = 3.66KK409 pKa = 10.11VIYY412 pKa = 10.68YY413 pKa = 9.89FDD415 pKa = 4.27KK416 pKa = 10.87EE417 pKa = 4.31QPQQMLANFFSNIEE431 pKa = 4.14TLPDD435 pKa = 3.77ISPQDD440 pKa = 3.48LSFLYY445 pKa = 10.93NLDD448 pKa = 4.71DD449 pKa = 5.05DD450 pKa = 5.12YY451 pKa = 11.89KK452 pKa = 11.19DD453 pKa = 3.3SKK455 pKa = 11.06LYY457 pKa = 10.69QCIKK461 pKa = 7.87THH463 pKa = 6.71AIEE466 pKa = 4.19VANDD470 pKa = 3.33NVKK473 pKa = 10.28HH474 pKa = 6.11KK475 pKa = 10.87KK476 pKa = 10.18EE477 pKa = 3.86NDD479 pKa = 3.38GLKK482 pKa = 9.93II483 pKa = 3.77
Molecular weight: 57.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1793 |
90 |
708 |
358.6 |
41.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.354 ± 1.119 | 1.171 ± 0.404 |
6.023 ± 0.635 | 4.964 ± 0.65 |
5.968 ± 0.864 | 4.518 ± 0.787 |
2.119 ± 0.467 | 6.247 ± 0.869 |
5.633 ± 1.045 | 9.314 ± 0.601 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.952 ± 0.343 | 8.31 ± 0.644 |
4.239 ± 0.811 | 5.02 ± 0.969 |
4.629 ± 0.288 | 7.808 ± 0.614 |
5.521 ± 0.681 | 4.462 ± 0.656 |
0.892 ± 0.15 | 5.856 ± 0.902 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
