
Neochlamydia sp. TUME1
Taxonomy: cellular organisms; Bacteria; PVC group; Chlamydiae; Chlamydiia; Parachlamydiales; Parachlamydiaceae; Neochlamydia; unclassified Neochlamydia
Average proteome isoelectric point is 7.3
Get precalculated fractions of proteins
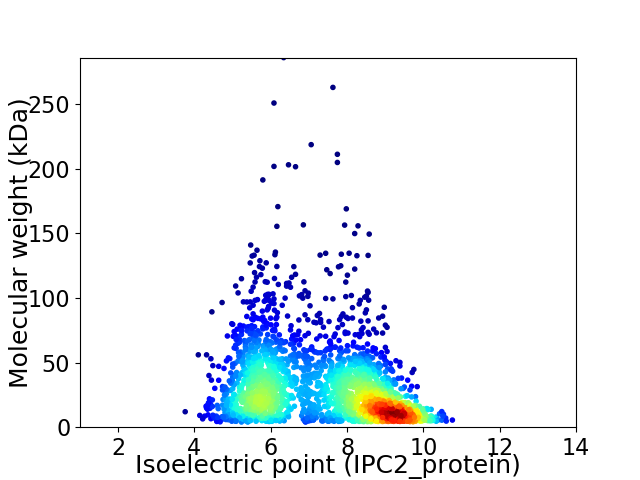
Virtual 2D-PAGE plot for 2344 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C1JL86|A0A0C1JL86_9BACT zf-IS66 domain-containing protein OS=Neochlamydia sp. TUME1 OX=1478174 GN=DB41_BK00010 PE=4 SV=1
DDD2 pKa = 4.76IAEEE6 pKa = 3.87RR7 pKa = 11.84KKK9 pKa = 10.11ITALDDD15 pKa = 4.18SRR17 pKa = 11.84IDDD20 pKa = 3.86TYYY23 pKa = 10.64PPEEE27 pKa = 4.3CQLSQLQTLDDD38 pKa = 3.43RR39 pKa = 11.84EEE41 pKa = 4.2QLTALPTEEE50 pKa = 4.7GQLSEEE56 pKa = 4.32QYYY59 pKa = 11.42YYY61 pKa = 11.24NQNQLTALPAEEE73 pKa = 4.49GRR75 pKa = 11.84LSEEE79 pKa = 4.12QSLEEE84 pKa = 4.53NQNQLTSLPAEEE96 pKa = 4.4GQLSKKK102 pKa = 11.21QVLHHH107 pKa = 7.09SQNQLTALPAEEE119 pKa = 4.63GQLSEEE125 pKa = 4.45QALDDD130 pKa = 4.36SQNQLTALPAEEE142 pKa = 4.63GQLSEEE148 pKa = 5.62KKK150 pKa = 9.97LDDD153 pKa = 3.74RR154 pKa = 11.84EEE156 pKa = 4.31EEE158 pKa = 4.51NNLPAEEE165 pKa = 4.46GRR167 pKa = 11.84LPQLQWLDDD176 pKa = 3.47SQNQLVSLPEEE187 pKa = 4.56IGRR190 pKa = 11.84LSQLQEEE197 pKa = 4.18NSSQNQLTNLPEEE210 pKa = 4.7EE211 pKa = 4.7GQLPQLQWLYYY222 pKa = 11.42NQNQLTALPSEEE234 pKa = 4.15RR235 pKa = 11.84QLFQLRR241 pKa = 11.84GLYYY245 pKa = 10.54NQNQLTSLPEEE256 pKa = 4.25EE257 pKa = 4.47GQLSQLQWLYYY268 pKa = 11.53NQNQLTSLPAEEE280 pKa = 4.62GQLPQLQEEE289 pKa = 4.18EEE291 pKa = 4.65NQNQLTSLPAEEE303 pKa = 4.54GQLSQLQVLYYY314 pKa = 11.07NQNQLASLPAEEE326 pKa = 4.5GQLPQLQEEE335 pKa = 4.35EEE337 pKa = 4.4SQNQLTSLPAEEE349 pKa = 4.4GQLSKKK355 pKa = 11.1QEEE358 pKa = 4.22YYY360 pKa = 11.14NQNQLTSLPVEEE372 pKa = 4.67GQLSQLQGFEEE383 pKa = 4.35SQNQLTALPVEEE395 pKa = 4.92GQLSQLQGLYYY406 pKa = 10.79NQNQLTSLPAEEE418 pKa = 4.54GQLSQLQEEE427 pKa = 4.04EEE429 pKa = 4.6NQNQLTSLPVDDD441 pKa = 3.53GQLSQLRR448 pKa = 11.84RR449 pKa = 11.84LEEE452 pKa = 4.49NHHH455 pKa = 6.22QLTALPTEEE464 pKa = 4.85GQLSQLQGLYYY475 pKa = 10.79NQNQLTSLPIEEE487 pKa = 4.61GQLSPLLQLEEE498 pKa = 4.29AE
DDD2 pKa = 4.76IAEEE6 pKa = 3.87RR7 pKa = 11.84KKK9 pKa = 10.11ITALDDD15 pKa = 4.18SRR17 pKa = 11.84IDDD20 pKa = 3.86TYYY23 pKa = 10.64PPEEE27 pKa = 4.3CQLSQLQTLDDD38 pKa = 3.43RR39 pKa = 11.84EEE41 pKa = 4.2QLTALPTEEE50 pKa = 4.7GQLSEEE56 pKa = 4.32QYYY59 pKa = 11.42YYY61 pKa = 11.24NQNQLTALPAEEE73 pKa = 4.49GRR75 pKa = 11.84LSEEE79 pKa = 4.12QSLEEE84 pKa = 4.53NQNQLTSLPAEEE96 pKa = 4.4GQLSKKK102 pKa = 11.21QVLHHH107 pKa = 7.09SQNQLTALPAEEE119 pKa = 4.63GQLSEEE125 pKa = 4.45QALDDD130 pKa = 4.36SQNQLTALPAEEE142 pKa = 4.63GQLSEEE148 pKa = 5.62KKK150 pKa = 9.97LDDD153 pKa = 3.74RR154 pKa = 11.84EEE156 pKa = 4.31EEE158 pKa = 4.51NNLPAEEE165 pKa = 4.46GRR167 pKa = 11.84LPQLQWLDDD176 pKa = 3.47SQNQLVSLPEEE187 pKa = 4.56IGRR190 pKa = 11.84LSQLQEEE197 pKa = 4.18NSSQNQLTNLPEEE210 pKa = 4.7EE211 pKa = 4.7GQLPQLQWLYYY222 pKa = 11.42NQNQLTALPSEEE234 pKa = 4.15RR235 pKa = 11.84QLFQLRR241 pKa = 11.84GLYYY245 pKa = 10.54NQNQLTSLPEEE256 pKa = 4.25EE257 pKa = 4.47GQLSQLQWLYYY268 pKa = 11.53NQNQLTSLPAEEE280 pKa = 4.62GQLPQLQEEE289 pKa = 4.18EEE291 pKa = 4.65NQNQLTSLPAEEE303 pKa = 4.54GQLSQLQVLYYY314 pKa = 11.07NQNQLASLPAEEE326 pKa = 4.5GQLPQLQEEE335 pKa = 4.35EEE337 pKa = 4.4SQNQLTSLPAEEE349 pKa = 4.4GQLSKKK355 pKa = 11.1QEEE358 pKa = 4.22YYY360 pKa = 11.14NQNQLTSLPVEEE372 pKa = 4.67GQLSQLQGFEEE383 pKa = 4.35SQNQLTALPVEEE395 pKa = 4.92GQLSQLQGLYYY406 pKa = 10.79NQNQLTSLPAEEE418 pKa = 4.54GQLSQLQEEE427 pKa = 4.04EEE429 pKa = 4.6NQNQLTSLPVDDD441 pKa = 3.53GQLSQLRR448 pKa = 11.84RR449 pKa = 11.84LEEE452 pKa = 4.49NHHH455 pKa = 6.22QLTALPTEEE464 pKa = 4.85GQLSQLQGLYYY475 pKa = 10.79NQNQLTSLPIEEE487 pKa = 4.61GQLSPLLQLEEE498 pKa = 4.29AE
Molecular weight: 56.02 kDa
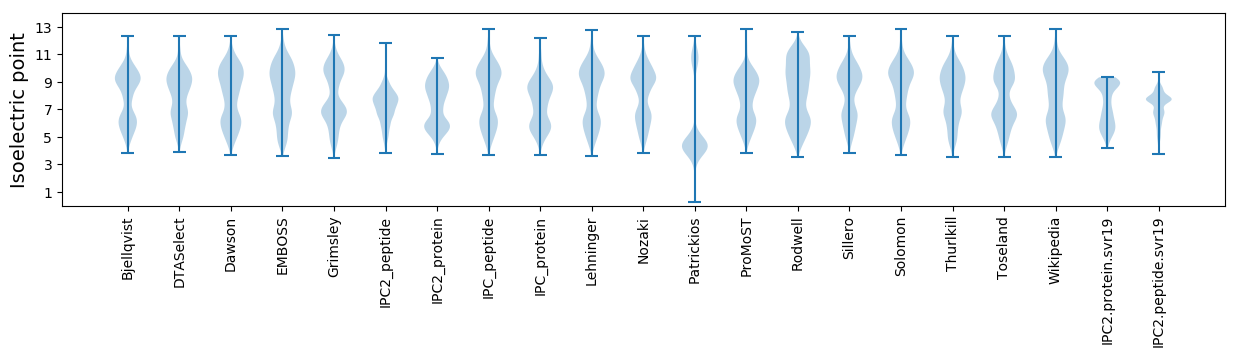
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C1HST5|A0A0C1HST5_9BACT Leucine-rich repeat-containing protein 1 OS=Neochlamydia sp. TUME1 OX=1478174 GN=lrrC1_1 PE=4 SV=1
MM1 pKa = 7.83IGRR4 pKa = 11.84KK5 pKa = 8.61LAYY8 pKa = 9.55HH9 pKa = 6.37KK10 pKa = 10.44HH11 pKa = 5.64HH12 pKa = 5.61LHH14 pKa = 6.72KK15 pKa = 10.73RR16 pKa = 11.84FLCSLVAKK24 pKa = 9.93RR25 pKa = 11.84SSGSLFNHH33 pKa = 6.44RR34 pKa = 11.84LDD36 pKa = 4.21YY37 pKa = 11.14SLSPYY42 pKa = 8.26LTKK45 pKa = 10.71SHH47 pKa = 6.82ASLGKK52 pKa = 9.6YY53 pKa = 10.0LSEE56 pKa = 4.2SFSKK60 pKa = 10.39FFVWLCHH67 pKa = 6.2ADD69 pKa = 3.71ALATLPHH76 pKa = 7.1PKK78 pKa = 9.94RR79 pKa = 11.84LLQRR83 pKa = 11.84NQRR86 pKa = 11.84LCLLDD91 pKa = 3.74ALRR94 pKa = 11.84LSFLISSCPSSLARR108 pKa = 11.84LQMRR112 pKa = 11.84VV113 pKa = 2.94
MM1 pKa = 7.83IGRR4 pKa = 11.84KK5 pKa = 8.61LAYY8 pKa = 9.55HH9 pKa = 6.37KK10 pKa = 10.44HH11 pKa = 5.64HH12 pKa = 5.61LHH14 pKa = 6.72KK15 pKa = 10.73RR16 pKa = 11.84FLCSLVAKK24 pKa = 9.93RR25 pKa = 11.84SSGSLFNHH33 pKa = 6.44RR34 pKa = 11.84LDD36 pKa = 4.21YY37 pKa = 11.14SLSPYY42 pKa = 8.26LTKK45 pKa = 10.71SHH47 pKa = 6.82ASLGKK52 pKa = 9.6YY53 pKa = 10.0LSEE56 pKa = 4.2SFSKK60 pKa = 10.39FFVWLCHH67 pKa = 6.2ADD69 pKa = 3.71ALATLPHH76 pKa = 7.1PKK78 pKa = 9.94RR79 pKa = 11.84LLQRR83 pKa = 11.84NQRR86 pKa = 11.84LCLLDD91 pKa = 3.74ALRR94 pKa = 11.84LSFLISSCPSSLARR108 pKa = 11.84LQMRR112 pKa = 11.84VV113 pKa = 2.94
Molecular weight: 13.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
675547 |
36 |
2477 |
288.2 |
32.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.218 ± 0.045 | 1.245 ± 0.019 |
4.08 ± 0.035 | 6.975 ± 0.056 |
4.413 ± 0.043 | 5.475 ± 0.055 |
2.566 ± 0.028 | 7.232 ± 0.047 |
7.395 ± 0.056 | 12.243 ± 0.099 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.105 ± 0.027 | 4.727 ± 0.041 |
4.156 ± 0.037 | 5.017 ± 0.057 |
4.278 ± 0.038 | 6.669 ± 0.046 |
4.697 ± 0.037 | 4.873 ± 0.052 |
1.106 ± 0.019 | 3.53 ± 0.036 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
