
Nitrospira sp. SCGC AG-212-E16
Taxonomy: cellular organisms; Bacteria; Nitrospirae; Nitrospira; Nitrospirales; Nitrospiraceae; Nitrospira; unclassified Nitrospira
Average proteome isoelectric point is 6.82
Get precalculated fractions of proteins
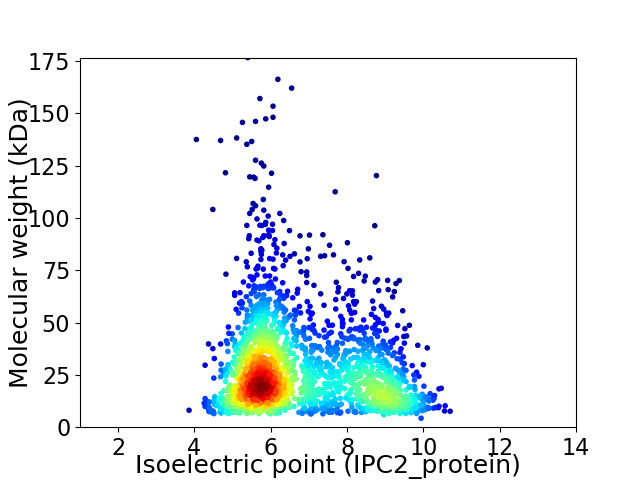
Virtual 2D-PAGE plot for 2093 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
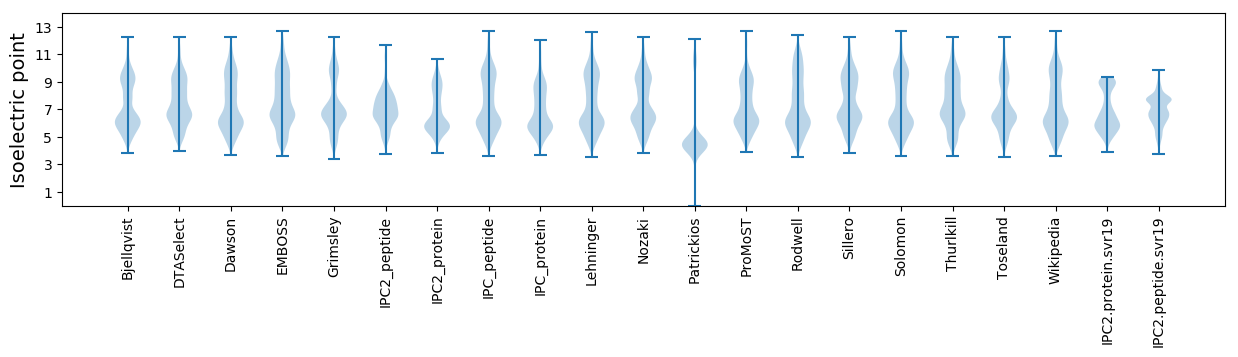
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A177QQH9|A0A177QQH9_9BACT Haemagg_act domain-containing protein OS=Nitrospira sp. SCGC AG-212-E16 OX=1799664 GN=AYO43_00010 PE=4 SV=1
MM1 pKa = 7.17GVVAEE6 pKa = 4.57GVVLLAVSAVACVACPSAMPDD27 pKa = 3.14TEE29 pKa = 4.22AAGGVWDD36 pKa = 4.32GVGGAVIVAMGGRR49 pKa = 11.84LVNVRR54 pKa = 11.84CCGVGIGAGDD64 pKa = 3.54IDD66 pKa = 4.59NEE68 pKa = 4.42GLCGSIGLAMCVGSVPEE85 pKa = 4.25GG86 pKa = 3.26
MM1 pKa = 7.17GVVAEE6 pKa = 4.57GVVLLAVSAVACVACPSAMPDD27 pKa = 3.14TEE29 pKa = 4.22AAGGVWDD36 pKa = 4.32GVGGAVIVAMGGRR49 pKa = 11.84LVNVRR54 pKa = 11.84CCGVGIGAGDD64 pKa = 3.54IDD66 pKa = 4.59NEE68 pKa = 4.42GLCGSIGLAMCVGSVPEE85 pKa = 4.25GG86 pKa = 3.26
Molecular weight: 8.09 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A177Q9V0|A0A177Q9V0_9BACT Uncharacterized protein OS=Nitrospira sp. SCGC AG-212-E16 OX=1799664 GN=AYO43_09030 PE=4 SV=1
MM1 pKa = 7.82RR2 pKa = 11.84LFALSIRR9 pKa = 11.84PLWHH13 pKa = 6.14GVCVPAGIMAQCHH26 pKa = 4.35TTFVALIAVSLILIAPHH43 pKa = 6.3QISAQQSTAPPSSLSTRR60 pKa = 11.84PLAPGTTEE68 pKa = 3.76SEE70 pKa = 4.18LRR72 pKa = 11.84PSCDD76 pKa = 2.84LCRR79 pKa = 11.84KK80 pKa = 8.88PEE82 pKa = 3.93SRR84 pKa = 11.84AGNTLNPHH92 pKa = 5.94RR93 pKa = 11.84LHH95 pKa = 6.83RR96 pKa = 11.84EE97 pKa = 3.92KK98 pKa = 10.86GLHH101 pKa = 5.03PHH103 pKa = 7.02KK104 pKa = 10.14KK105 pKa = 7.57PKK107 pKa = 10.01GMRR110 pKa = 11.84RR111 pKa = 11.84SSRR114 pKa = 11.84RR115 pKa = 11.84SVRR118 pKa = 11.84SLLRR122 pKa = 11.84RR123 pKa = 11.84QAFAPLGGIEE133 pKa = 4.04RR134 pKa = 11.84TVDD137 pKa = 2.88SRR139 pKa = 11.84QVHH142 pKa = 5.4VVDD145 pKa = 4.73GDD147 pKa = 3.62TFRR150 pKa = 11.84YY151 pKa = 7.28GTEE154 pKa = 3.73RR155 pKa = 11.84VRR157 pKa = 11.84LRR159 pKa = 11.84GIDD162 pKa = 3.62TPEE165 pKa = 4.42LDD167 pKa = 4.21EE168 pKa = 5.49PNGQAARR175 pKa = 11.84LRR177 pKa = 11.84LEE179 pKa = 4.28EE180 pKa = 4.73LLHH183 pKa = 6.02SGQVRR188 pKa = 11.84IVPHH192 pKa = 6.1GRR194 pKa = 11.84DD195 pKa = 3.26VYY197 pKa = 11.34DD198 pKa = 3.84RR199 pKa = 11.84LVADD203 pKa = 3.65VFVDD207 pKa = 3.68GRR209 pKa = 11.84NVTDD213 pKa = 3.62MLTQEE218 pKa = 5.62GYY220 pKa = 11.11AKK222 pKa = 10.23PRR224 pKa = 11.84SS225 pKa = 3.66
MM1 pKa = 7.82RR2 pKa = 11.84LFALSIRR9 pKa = 11.84PLWHH13 pKa = 6.14GVCVPAGIMAQCHH26 pKa = 4.35TTFVALIAVSLILIAPHH43 pKa = 6.3QISAQQSTAPPSSLSTRR60 pKa = 11.84PLAPGTTEE68 pKa = 3.76SEE70 pKa = 4.18LRR72 pKa = 11.84PSCDD76 pKa = 2.84LCRR79 pKa = 11.84KK80 pKa = 8.88PEE82 pKa = 3.93SRR84 pKa = 11.84AGNTLNPHH92 pKa = 5.94RR93 pKa = 11.84LHH95 pKa = 6.83RR96 pKa = 11.84EE97 pKa = 3.92KK98 pKa = 10.86GLHH101 pKa = 5.03PHH103 pKa = 7.02KK104 pKa = 10.14KK105 pKa = 7.57PKK107 pKa = 10.01GMRR110 pKa = 11.84RR111 pKa = 11.84SSRR114 pKa = 11.84RR115 pKa = 11.84SVRR118 pKa = 11.84SLLRR122 pKa = 11.84RR123 pKa = 11.84QAFAPLGGIEE133 pKa = 4.04RR134 pKa = 11.84TVDD137 pKa = 2.88SRR139 pKa = 11.84QVHH142 pKa = 5.4VVDD145 pKa = 4.73GDD147 pKa = 3.62TFRR150 pKa = 11.84YY151 pKa = 7.28GTEE154 pKa = 3.73RR155 pKa = 11.84VRR157 pKa = 11.84LRR159 pKa = 11.84GIDD162 pKa = 3.62TPEE165 pKa = 4.42LDD167 pKa = 4.21EE168 pKa = 5.49PNGQAARR175 pKa = 11.84LRR177 pKa = 11.84LEE179 pKa = 4.28EE180 pKa = 4.73LLHH183 pKa = 6.02SGQVRR188 pKa = 11.84IVPHH192 pKa = 6.1GRR194 pKa = 11.84DD195 pKa = 3.26VYY197 pKa = 11.34DD198 pKa = 3.84RR199 pKa = 11.84LVADD203 pKa = 3.65VFVDD207 pKa = 3.68GRR209 pKa = 11.84NVTDD213 pKa = 3.62MLTQEE218 pKa = 5.62GYY220 pKa = 11.11AKK222 pKa = 10.23PRR224 pKa = 11.84SS225 pKa = 3.66
Molecular weight: 25.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
575471 |
37 |
1567 |
275.0 |
30.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.556 ± 0.057 | 1.015 ± 0.022 |
5.097 ± 0.035 | 6.047 ± 0.047 |
3.657 ± 0.034 | 7.8 ± 0.056 |
2.401 ± 0.031 | 5.363 ± 0.045 |
4.358 ± 0.057 | 10.474 ± 0.064 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.477 ± 0.027 | 2.724 ± 0.034 |
5.009 ± 0.039 | 4.015 ± 0.035 |
6.653 ± 0.06 | 5.897 ± 0.049 |
5.819 ± 0.042 | 7.688 ± 0.053 |
1.367 ± 0.023 | 2.583 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
