
Rat associated porprismacovirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Porprismacovirus
Average proteome isoelectric point is 5.92
Get precalculated fractions of proteins
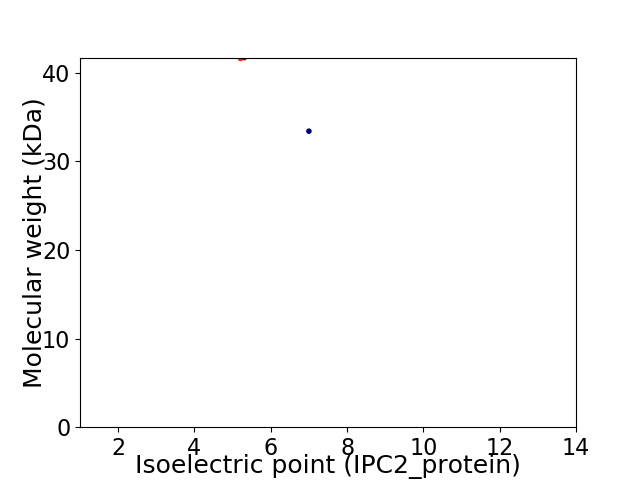
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0K1R815|A0A0K1R815_9VIRU Rep protein OS=Rat associated porprismacovirus 1 OX=2170126 PE=4 SV=1
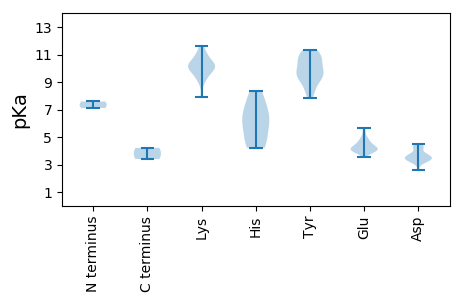
MM1 pKa = 7.11VSASISEE8 pKa = 4.34IYY10 pKa = 10.45DD11 pKa = 3.34LSTTVGQGTVLKK23 pKa = 10.43IHH25 pKa = 6.62TPTGNNVRR33 pKa = 11.84RR34 pKa = 11.84HH35 pKa = 4.83LLGYY39 pKa = 9.1FLQYY43 pKa = 10.96KK44 pKa = 9.06KK45 pKa = 10.68FRR47 pKa = 11.84YY48 pKa = 9.41VGAKK52 pKa = 7.92VTLVPASTLPADD64 pKa = 4.26PLQLSYY70 pKa = 11.3AAGEE74 pKa = 4.04PTIDD78 pKa = 3.86PRR80 pKa = 11.84DD81 pKa = 3.72MVNPILWRR89 pKa = 11.84HH90 pKa = 4.22YY91 pKa = 10.22HH92 pKa = 5.89GEE94 pKa = 4.01AMLTDD99 pKa = 4.15PLARR103 pKa = 11.84TFPEE107 pKa = 4.78PYY109 pKa = 9.91GSEE112 pKa = 4.06INPGTSEE119 pKa = 4.77IIGTSIDD126 pKa = 3.63RR127 pKa = 11.84QDD129 pKa = 3.71YY130 pKa = 9.1TSGAGDD136 pKa = 3.43INQIYY141 pKa = 9.12PMCLMDD147 pKa = 4.41PSFRR151 pKa = 11.84KK152 pKa = 10.28AGVQSGFSTFVKK164 pKa = 10.0PYY166 pKa = 9.64VYY168 pKa = 10.89KK169 pKa = 10.93LATNAQLLPMRR180 pKa = 11.84GFTTPSSGAAYY191 pKa = 7.84TWNTNKK197 pKa = 9.69IEE199 pKa = 3.99PTIGGTQEE207 pKa = 4.17SYY209 pKa = 11.2LVDD212 pKa = 3.66PVLSEE217 pKa = 5.0DD218 pKa = 3.46NSSYY222 pKa = 11.31VPDD225 pKa = 4.3DD226 pKa = 3.61LKK228 pKa = 11.64GNVIFTNKK236 pKa = 9.65LDD238 pKa = 3.74RR239 pKa = 11.84LGWMDD244 pKa = 3.59TVTRR248 pKa = 11.84TLLTGSKK255 pKa = 9.2PLAVSPIPGPASSGTAIEE273 pKa = 4.78SGTGGNQTGGNFALQTYY290 pKa = 9.19LPNIPMLYY298 pKa = 10.87VMMPPAYY305 pKa = 9.43KK306 pKa = 10.24SLFYY310 pKa = 10.55FRR312 pKa = 11.84LIIKK316 pKa = 9.97HH317 pKa = 4.94YY318 pKa = 10.59FEE320 pKa = 5.4FGGFRR325 pKa = 11.84SCLNVQSYY333 pKa = 10.76GSASVYY339 pKa = 9.72VAQPLNPVTSSNVSEE354 pKa = 4.22IASMVSGIADD364 pKa = 4.46GGPDD368 pKa = 3.38SLEE371 pKa = 4.04VEE373 pKa = 4.12NGEE376 pKa = 4.33IVSSADD382 pKa = 3.1GVLL385 pKa = 3.4
MM1 pKa = 7.11VSASISEE8 pKa = 4.34IYY10 pKa = 10.45DD11 pKa = 3.34LSTTVGQGTVLKK23 pKa = 10.43IHH25 pKa = 6.62TPTGNNVRR33 pKa = 11.84RR34 pKa = 11.84HH35 pKa = 4.83LLGYY39 pKa = 9.1FLQYY43 pKa = 10.96KK44 pKa = 9.06KK45 pKa = 10.68FRR47 pKa = 11.84YY48 pKa = 9.41VGAKK52 pKa = 7.92VTLVPASTLPADD64 pKa = 4.26PLQLSYY70 pKa = 11.3AAGEE74 pKa = 4.04PTIDD78 pKa = 3.86PRR80 pKa = 11.84DD81 pKa = 3.72MVNPILWRR89 pKa = 11.84HH90 pKa = 4.22YY91 pKa = 10.22HH92 pKa = 5.89GEE94 pKa = 4.01AMLTDD99 pKa = 4.15PLARR103 pKa = 11.84TFPEE107 pKa = 4.78PYY109 pKa = 9.91GSEE112 pKa = 4.06INPGTSEE119 pKa = 4.77IIGTSIDD126 pKa = 3.63RR127 pKa = 11.84QDD129 pKa = 3.71YY130 pKa = 9.1TSGAGDD136 pKa = 3.43INQIYY141 pKa = 9.12PMCLMDD147 pKa = 4.41PSFRR151 pKa = 11.84KK152 pKa = 10.28AGVQSGFSTFVKK164 pKa = 10.0PYY166 pKa = 9.64VYY168 pKa = 10.89KK169 pKa = 10.93LATNAQLLPMRR180 pKa = 11.84GFTTPSSGAAYY191 pKa = 7.84TWNTNKK197 pKa = 9.69IEE199 pKa = 3.99PTIGGTQEE207 pKa = 4.17SYY209 pKa = 11.2LVDD212 pKa = 3.66PVLSEE217 pKa = 5.0DD218 pKa = 3.46NSSYY222 pKa = 11.31VPDD225 pKa = 4.3DD226 pKa = 3.61LKK228 pKa = 11.64GNVIFTNKK236 pKa = 9.65LDD238 pKa = 3.74RR239 pKa = 11.84LGWMDD244 pKa = 3.59TVTRR248 pKa = 11.84TLLTGSKK255 pKa = 9.2PLAVSPIPGPASSGTAIEE273 pKa = 4.78SGTGGNQTGGNFALQTYY290 pKa = 9.19LPNIPMLYY298 pKa = 10.87VMMPPAYY305 pKa = 9.43KK306 pKa = 10.24SLFYY310 pKa = 10.55FRR312 pKa = 11.84LIIKK316 pKa = 9.97HH317 pKa = 4.94YY318 pKa = 10.59FEE320 pKa = 5.4FGGFRR325 pKa = 11.84SCLNVQSYY333 pKa = 10.76GSASVYY339 pKa = 9.72VAQPLNPVTSSNVSEE354 pKa = 4.22IASMVSGIADD364 pKa = 4.46GGPDD368 pKa = 3.38SLEE371 pKa = 4.04VEE373 pKa = 4.12NGEE376 pKa = 4.33IVSSADD382 pKa = 3.1GVLL385 pKa = 3.4
Molecular weight: 41.63 kDa
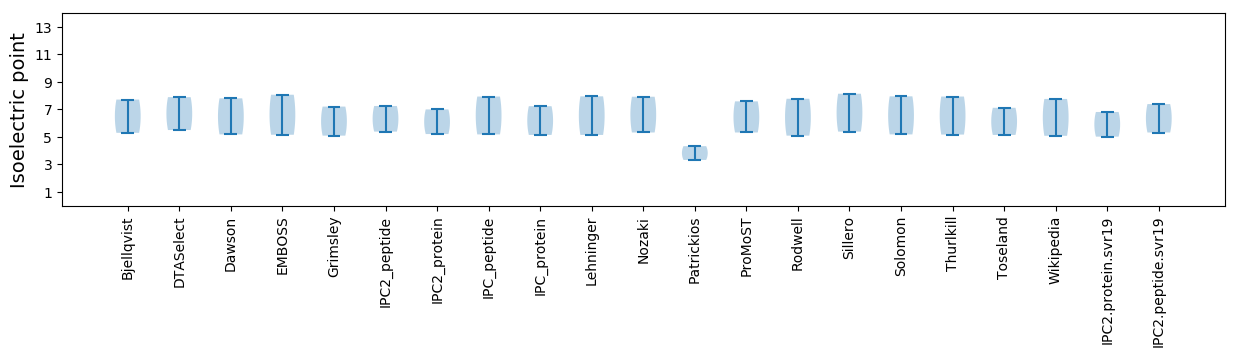
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0K1R815|A0A0K1R815_9VIRU Rep protein OS=Rat associated porprismacovirus 1 OX=2170126 PE=4 SV=1
MM1 pKa = 7.59RR2 pKa = 11.84HH3 pKa = 5.85PVSMTKK9 pKa = 10.21CYY11 pKa = 10.01MITAPRR17 pKa = 11.84DD18 pKa = 3.3EE19 pKa = 4.44EE20 pKa = 4.46MYY22 pKa = 11.12ALRR25 pKa = 11.84IWRR28 pKa = 11.84YY29 pKa = 8.42INSHH33 pKa = 6.59DD34 pKa = 3.52VHH36 pKa = 7.73KK37 pKa = 10.43WIIASEE43 pKa = 3.96IGRR46 pKa = 11.84NGYY49 pKa = 8.25KK50 pKa = 9.71HH51 pKa = 4.23WQIRR55 pKa = 11.84IKK57 pKa = 10.15TSDD60 pKa = 3.39PDD62 pKa = 3.55FFSFEE67 pKa = 4.1EE68 pKa = 4.38RR69 pKa = 11.84EE70 pKa = 4.19EE71 pKa = 4.64PYY73 pKa = 9.79LTANWKK79 pKa = 9.56IEE81 pKa = 3.79YY82 pKa = 9.5RR83 pKa = 11.84KK84 pKa = 10.52VKK86 pKa = 10.31IGTGWANINIPRR98 pKa = 11.84SHH100 pKa = 8.37VEE102 pKa = 3.87EE103 pKa = 5.63CSDD106 pKa = 3.31DD107 pKa = 3.13WDD109 pKa = 4.27YY110 pKa = 10.25EE111 pKa = 4.59TKK113 pKa = 10.24EE114 pKa = 3.96GRR116 pKa = 11.84YY117 pKa = 8.71LASWDD122 pKa = 3.68TPEE125 pKa = 3.88VRR127 pKa = 11.84KK128 pKa = 10.0LRR130 pKa = 11.84FGQPRR135 pKa = 11.84WHH137 pKa = 6.37QEE139 pKa = 3.58AIINRR144 pKa = 11.84LEE146 pKa = 4.04STNDD150 pKa = 3.16RR151 pKa = 11.84EE152 pKa = 4.44VMVWYY157 pKa = 10.01DD158 pKa = 3.28PTGNSGKK165 pKa = 9.87SWLVGHH171 pKa = 7.46LYY173 pKa = 8.36EE174 pKa = 5.03TGQAYY179 pKa = 9.22YY180 pKa = 10.43LPPTMTSVQSMLQMMASLAIQDD202 pKa = 4.47RR203 pKa = 11.84EE204 pKa = 4.22DD205 pKa = 3.28GRR207 pKa = 11.84PPRR210 pKa = 11.84RR211 pKa = 11.84YY212 pKa = 9.8VVIDD216 pKa = 3.92IPRR219 pKa = 11.84TWKK222 pKa = 9.1WSKK225 pKa = 11.05DD226 pKa = 3.39LYY228 pKa = 10.91CAIEE232 pKa = 4.27TIKK235 pKa = 10.98DD236 pKa = 3.56GLIVDD241 pKa = 4.29PRR243 pKa = 11.84YY244 pKa = 9.57SARR247 pKa = 11.84PINIRR252 pKa = 11.84GVKK255 pKa = 10.05VLVLSNDD262 pKa = 3.35RR263 pKa = 11.84PSLDD267 pKa = 3.47KK268 pKa = 11.28LSVDD272 pKa = 2.63RR273 pKa = 11.84WVVEE277 pKa = 4.09NTPMLL282 pKa = 4.21
MM1 pKa = 7.59RR2 pKa = 11.84HH3 pKa = 5.85PVSMTKK9 pKa = 10.21CYY11 pKa = 10.01MITAPRR17 pKa = 11.84DD18 pKa = 3.3EE19 pKa = 4.44EE20 pKa = 4.46MYY22 pKa = 11.12ALRR25 pKa = 11.84IWRR28 pKa = 11.84YY29 pKa = 8.42INSHH33 pKa = 6.59DD34 pKa = 3.52VHH36 pKa = 7.73KK37 pKa = 10.43WIIASEE43 pKa = 3.96IGRR46 pKa = 11.84NGYY49 pKa = 8.25KK50 pKa = 9.71HH51 pKa = 4.23WQIRR55 pKa = 11.84IKK57 pKa = 10.15TSDD60 pKa = 3.39PDD62 pKa = 3.55FFSFEE67 pKa = 4.1EE68 pKa = 4.38RR69 pKa = 11.84EE70 pKa = 4.19EE71 pKa = 4.64PYY73 pKa = 9.79LTANWKK79 pKa = 9.56IEE81 pKa = 3.79YY82 pKa = 9.5RR83 pKa = 11.84KK84 pKa = 10.52VKK86 pKa = 10.31IGTGWANINIPRR98 pKa = 11.84SHH100 pKa = 8.37VEE102 pKa = 3.87EE103 pKa = 5.63CSDD106 pKa = 3.31DD107 pKa = 3.13WDD109 pKa = 4.27YY110 pKa = 10.25EE111 pKa = 4.59TKK113 pKa = 10.24EE114 pKa = 3.96GRR116 pKa = 11.84YY117 pKa = 8.71LASWDD122 pKa = 3.68TPEE125 pKa = 3.88VRR127 pKa = 11.84KK128 pKa = 10.0LRR130 pKa = 11.84FGQPRR135 pKa = 11.84WHH137 pKa = 6.37QEE139 pKa = 3.58AIINRR144 pKa = 11.84LEE146 pKa = 4.04STNDD150 pKa = 3.16RR151 pKa = 11.84EE152 pKa = 4.44VMVWYY157 pKa = 10.01DD158 pKa = 3.28PTGNSGKK165 pKa = 9.87SWLVGHH171 pKa = 7.46LYY173 pKa = 8.36EE174 pKa = 5.03TGQAYY179 pKa = 9.22YY180 pKa = 10.43LPPTMTSVQSMLQMMASLAIQDD202 pKa = 4.47RR203 pKa = 11.84EE204 pKa = 4.22DD205 pKa = 3.28GRR207 pKa = 11.84PPRR210 pKa = 11.84RR211 pKa = 11.84YY212 pKa = 9.8VVIDD216 pKa = 3.92IPRR219 pKa = 11.84TWKK222 pKa = 9.1WSKK225 pKa = 11.05DD226 pKa = 3.39LYY228 pKa = 10.91CAIEE232 pKa = 4.27TIKK235 pKa = 10.98DD236 pKa = 3.56GLIVDD241 pKa = 4.29PRR243 pKa = 11.84YY244 pKa = 9.57SARR247 pKa = 11.84PINIRR252 pKa = 11.84GVKK255 pKa = 10.05VLVLSNDD262 pKa = 3.35RR263 pKa = 11.84PSLDD267 pKa = 3.47KK268 pKa = 11.28LSVDD272 pKa = 2.63RR273 pKa = 11.84WVVEE277 pKa = 4.09NTPMLL282 pKa = 4.21
Molecular weight: 33.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
667 |
282 |
385 |
333.5 |
37.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.397 ± 0.77 | 0.75 ± 0.212 |
5.547 ± 0.803 | 5.247 ± 1.244 |
2.699 ± 0.863 | 7.346 ± 1.845 |
1.799 ± 0.461 | 6.597 ± 0.812 |
4.198 ± 0.756 | 7.496 ± 0.99 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.148 ± 0.268 | 4.348 ± 0.301 |
7.046 ± 0.686 | 2.849 ± 0.247 |
5.697 ± 2.136 | 8.396 ± 1.118 |
6.897 ± 1.063 | 6.747 ± 0.245 |
2.399 ± 1.491 | 5.397 ± 0.053 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
