
Changjiang tombus-like virus 17
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.26
Get precalculated fractions of proteins
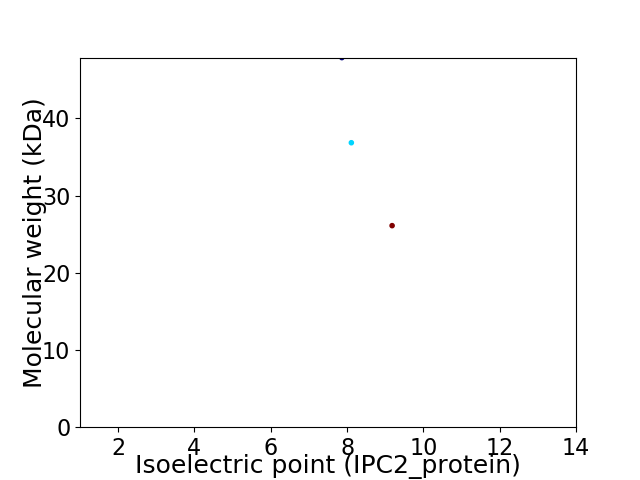
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KFU6|A0A1L3KFU6_9VIRU RNA-directed RNA polymerase OS=Changjiang tombus-like virus 17 OX=1922810 PE=4 SV=1
MM1 pKa = 6.95YY2 pKa = 10.19RR3 pKa = 11.84GFRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84YY10 pKa = 8.82EE11 pKa = 3.52QAKK14 pKa = 9.94QNLSISGPLIDD25 pKa = 4.99SRR27 pKa = 11.84VKK29 pKa = 10.34MFVKK33 pKa = 10.04MEE35 pKa = 4.56GIKK38 pKa = 10.61FSSEE42 pKa = 4.17KK43 pKa = 10.46INPSCRR49 pKa = 11.84AIQFRR54 pKa = 11.84SPEE57 pKa = 3.88YY58 pKa = 9.87QLQVACFIKK67 pKa = 10.49PIEE70 pKa = 4.26HH71 pKa = 7.32LLYY74 pKa = 10.4CSSGPYY80 pKa = 9.55PYY82 pKa = 9.8PASQFIAKK90 pKa = 9.36NLNGVEE96 pKa = 4.02RR97 pKa = 11.84AQLLWEE103 pKa = 4.96KK104 pKa = 10.4YY105 pKa = 10.48ANLPGCEE112 pKa = 3.91ILMLDD117 pKa = 3.9ASRR120 pKa = 11.84FDD122 pKa = 3.38GHH124 pKa = 5.07VTPRR128 pKa = 11.84LNSIEE133 pKa = 3.97EE134 pKa = 4.04HH135 pKa = 7.04FYY137 pKa = 8.6TTICPDD143 pKa = 3.29PFFRR147 pKa = 11.84SLLKK151 pKa = 10.36ARR153 pKa = 11.84KK154 pKa = 8.72VNKK157 pKa = 10.04GSAKK161 pKa = 10.3GADD164 pKa = 3.66FSVSYY169 pKa = 10.01SLKK172 pKa = 8.99GGRR175 pKa = 11.84MSGDD179 pKa = 3.2MDD181 pKa = 4.05TASSNCLLMSSMLALLGSTSCLSYY205 pKa = 11.42DD206 pKa = 3.84FLVDD210 pKa = 3.33GDD212 pKa = 4.49DD213 pKa = 4.08SVFFYY218 pKa = 11.03LGDD221 pKa = 3.78TLTDD225 pKa = 3.34DD226 pKa = 4.43HH227 pKa = 8.01IKK229 pKa = 10.8EE230 pKa = 4.24FFLEE234 pKa = 4.69CGMEE238 pKa = 3.89MKK240 pKa = 9.81IDD242 pKa = 3.25KK243 pKa = 8.43RR244 pKa = 11.84TRR246 pKa = 11.84DD247 pKa = 3.33FWSLDD252 pKa = 3.34FCQGKK257 pKa = 8.47PVSLPGGISLVRR269 pKa = 11.84DD270 pKa = 4.18PIKK273 pKa = 11.08VMSKK277 pKa = 10.76AGINDD282 pKa = 3.63KK283 pKa = 10.9FSDD286 pKa = 3.56PLLRR290 pKa = 11.84CRR292 pKa = 11.84ILEE295 pKa = 4.33VICLGEE301 pKa = 3.87LSLVKK306 pKa = 10.42GCPILQHH313 pKa = 6.3FFDD316 pKa = 4.49RR317 pKa = 11.84LLYY320 pKa = 9.35IARR323 pKa = 11.84RR324 pKa = 11.84SLGKK328 pKa = 10.23KK329 pKa = 8.97KK330 pKa = 10.09RR331 pKa = 11.84KK332 pKa = 7.83KK333 pKa = 10.72LKK335 pKa = 9.44MEE337 pKa = 3.69WMSYY341 pKa = 6.9RR342 pKa = 11.84QRR344 pKa = 11.84NEE346 pKa = 3.93FRR348 pKa = 11.84GEE350 pKa = 3.08WWKK353 pKa = 11.3RR354 pKa = 11.84EE355 pKa = 3.91VLPITPEE362 pKa = 3.59ARR364 pKa = 11.84EE365 pKa = 4.04TLSRR369 pKa = 11.84AWGISIEE376 pKa = 4.22EE377 pKa = 3.93QLSLEE382 pKa = 4.08KK383 pKa = 10.87QIDD386 pKa = 3.96GFDD389 pKa = 4.45FDD391 pKa = 5.18PMSTPIRR398 pKa = 11.84GEE400 pKa = 4.0GVDD403 pKa = 3.56VSNWLFDD410 pKa = 3.33AFTRR414 pKa = 11.84EE415 pKa = 4.26SFF417 pKa = 3.4
MM1 pKa = 6.95YY2 pKa = 10.19RR3 pKa = 11.84GFRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84YY10 pKa = 8.82EE11 pKa = 3.52QAKK14 pKa = 9.94QNLSISGPLIDD25 pKa = 4.99SRR27 pKa = 11.84VKK29 pKa = 10.34MFVKK33 pKa = 10.04MEE35 pKa = 4.56GIKK38 pKa = 10.61FSSEE42 pKa = 4.17KK43 pKa = 10.46INPSCRR49 pKa = 11.84AIQFRR54 pKa = 11.84SPEE57 pKa = 3.88YY58 pKa = 9.87QLQVACFIKK67 pKa = 10.49PIEE70 pKa = 4.26HH71 pKa = 7.32LLYY74 pKa = 10.4CSSGPYY80 pKa = 9.55PYY82 pKa = 9.8PASQFIAKK90 pKa = 9.36NLNGVEE96 pKa = 4.02RR97 pKa = 11.84AQLLWEE103 pKa = 4.96KK104 pKa = 10.4YY105 pKa = 10.48ANLPGCEE112 pKa = 3.91ILMLDD117 pKa = 3.9ASRR120 pKa = 11.84FDD122 pKa = 3.38GHH124 pKa = 5.07VTPRR128 pKa = 11.84LNSIEE133 pKa = 3.97EE134 pKa = 4.04HH135 pKa = 7.04FYY137 pKa = 8.6TTICPDD143 pKa = 3.29PFFRR147 pKa = 11.84SLLKK151 pKa = 10.36ARR153 pKa = 11.84KK154 pKa = 8.72VNKK157 pKa = 10.04GSAKK161 pKa = 10.3GADD164 pKa = 3.66FSVSYY169 pKa = 10.01SLKK172 pKa = 8.99GGRR175 pKa = 11.84MSGDD179 pKa = 3.2MDD181 pKa = 4.05TASSNCLLMSSMLALLGSTSCLSYY205 pKa = 11.42DD206 pKa = 3.84FLVDD210 pKa = 3.33GDD212 pKa = 4.49DD213 pKa = 4.08SVFFYY218 pKa = 11.03LGDD221 pKa = 3.78TLTDD225 pKa = 3.34DD226 pKa = 4.43HH227 pKa = 8.01IKK229 pKa = 10.8EE230 pKa = 4.24FFLEE234 pKa = 4.69CGMEE238 pKa = 3.89MKK240 pKa = 9.81IDD242 pKa = 3.25KK243 pKa = 8.43RR244 pKa = 11.84TRR246 pKa = 11.84DD247 pKa = 3.33FWSLDD252 pKa = 3.34FCQGKK257 pKa = 8.47PVSLPGGISLVRR269 pKa = 11.84DD270 pKa = 4.18PIKK273 pKa = 11.08VMSKK277 pKa = 10.76AGINDD282 pKa = 3.63KK283 pKa = 10.9FSDD286 pKa = 3.56PLLRR290 pKa = 11.84CRR292 pKa = 11.84ILEE295 pKa = 4.33VICLGEE301 pKa = 3.87LSLVKK306 pKa = 10.42GCPILQHH313 pKa = 6.3FFDD316 pKa = 4.49RR317 pKa = 11.84LLYY320 pKa = 9.35IARR323 pKa = 11.84RR324 pKa = 11.84SLGKK328 pKa = 10.23KK329 pKa = 8.97KK330 pKa = 10.09RR331 pKa = 11.84KK332 pKa = 7.83KK333 pKa = 10.72LKK335 pKa = 9.44MEE337 pKa = 3.69WMSYY341 pKa = 6.9RR342 pKa = 11.84QRR344 pKa = 11.84NEE346 pKa = 3.93FRR348 pKa = 11.84GEE350 pKa = 3.08WWKK353 pKa = 11.3RR354 pKa = 11.84EE355 pKa = 3.91VLPITPEE362 pKa = 3.59ARR364 pKa = 11.84EE365 pKa = 4.04TLSRR369 pKa = 11.84AWGISIEE376 pKa = 4.22EE377 pKa = 3.93QLSLEE382 pKa = 4.08KK383 pKa = 10.87QIDD386 pKa = 3.96GFDD389 pKa = 4.45FDD391 pKa = 5.18PMSTPIRR398 pKa = 11.84GEE400 pKa = 4.0GVDD403 pKa = 3.56VSNWLFDD410 pKa = 3.33AFTRR414 pKa = 11.84EE415 pKa = 4.26SFF417 pKa = 3.4
Molecular weight: 47.89 kDa
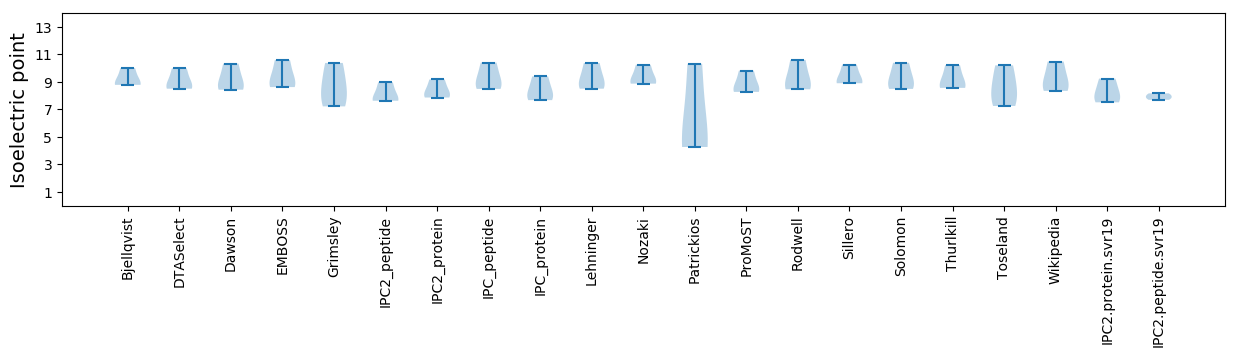
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KFU6|A0A1L3KFU6_9VIRU RNA-directed RNA polymerase OS=Changjiang tombus-like virus 17 OX=1922810 PE=4 SV=1
MM1 pKa = 7.88RR2 pKa = 11.84FLTNVPSVYY11 pKa = 10.31EE12 pKa = 4.19GFSGRR17 pKa = 11.84LRR19 pKa = 11.84SWITEE24 pKa = 3.45NRR26 pKa = 11.84RR27 pKa = 11.84NIILGSFCLIASGLTFRR44 pKa = 11.84KK45 pKa = 8.39TSQLTLLFRR54 pKa = 11.84SLLGLLGAGLSFTLGNNLTNLVSLQLEE81 pKa = 4.34NKK83 pKa = 10.09AKK85 pKa = 9.57IARR88 pKa = 11.84RR89 pKa = 11.84GTRR92 pKa = 11.84GKK94 pKa = 9.31MRR96 pKa = 11.84AIRR99 pKa = 11.84HH100 pKa = 5.27RR101 pKa = 11.84RR102 pKa = 11.84VPLTNLHH109 pKa = 6.51FEE111 pKa = 4.28VEE113 pKa = 4.53SLCFDD118 pKa = 4.14GLRR121 pKa = 11.84NSSDD125 pKa = 3.15VTDD128 pKa = 3.65IARR131 pKa = 11.84RR132 pKa = 11.84LKK134 pKa = 10.23QIAMSSGLEE143 pKa = 4.16VYY145 pKa = 10.94SDD147 pKa = 3.98DD148 pKa = 3.53GTVNKK153 pKa = 9.95VPVKK157 pKa = 9.78DD158 pKa = 4.12LNMALVDD165 pKa = 4.1SVVRR169 pKa = 11.84DD170 pKa = 4.02CVLHH174 pKa = 7.75ISDD177 pKa = 5.08PDD179 pKa = 3.51VLRR182 pKa = 11.84DD183 pKa = 3.57IEE185 pKa = 4.18VDD187 pKa = 3.16AEE189 pKa = 4.05NRR191 pKa = 11.84DD192 pKa = 3.32MAMRR196 pKa = 11.84RR197 pKa = 11.84LLSLGNAGQILSGWEE212 pKa = 3.76HH213 pKa = 6.44LKK215 pKa = 10.57ISALKK220 pKa = 10.34CGAAILGRR228 pKa = 11.84SYY230 pKa = 11.41LVGRR234 pKa = 11.84NN235 pKa = 3.28
MM1 pKa = 7.88RR2 pKa = 11.84FLTNVPSVYY11 pKa = 10.31EE12 pKa = 4.19GFSGRR17 pKa = 11.84LRR19 pKa = 11.84SWITEE24 pKa = 3.45NRR26 pKa = 11.84RR27 pKa = 11.84NIILGSFCLIASGLTFRR44 pKa = 11.84KK45 pKa = 8.39TSQLTLLFRR54 pKa = 11.84SLLGLLGAGLSFTLGNNLTNLVSLQLEE81 pKa = 4.34NKK83 pKa = 10.09AKK85 pKa = 9.57IARR88 pKa = 11.84RR89 pKa = 11.84GTRR92 pKa = 11.84GKK94 pKa = 9.31MRR96 pKa = 11.84AIRR99 pKa = 11.84HH100 pKa = 5.27RR101 pKa = 11.84RR102 pKa = 11.84VPLTNLHH109 pKa = 6.51FEE111 pKa = 4.28VEE113 pKa = 4.53SLCFDD118 pKa = 4.14GLRR121 pKa = 11.84NSSDD125 pKa = 3.15VTDD128 pKa = 3.65IARR131 pKa = 11.84RR132 pKa = 11.84LKK134 pKa = 10.23QIAMSSGLEE143 pKa = 4.16VYY145 pKa = 10.94SDD147 pKa = 3.98DD148 pKa = 3.53GTVNKK153 pKa = 9.95VPVKK157 pKa = 9.78DD158 pKa = 4.12LNMALVDD165 pKa = 4.1SVVRR169 pKa = 11.84DD170 pKa = 4.02CVLHH174 pKa = 7.75ISDD177 pKa = 5.08PDD179 pKa = 3.51VLRR182 pKa = 11.84DD183 pKa = 3.57IEE185 pKa = 4.18VDD187 pKa = 3.16AEE189 pKa = 4.05NRR191 pKa = 11.84DD192 pKa = 3.32MAMRR196 pKa = 11.84RR197 pKa = 11.84LLSLGNAGQILSGWEE212 pKa = 3.76HH213 pKa = 6.44LKK215 pKa = 10.57ISALKK220 pKa = 10.34CGAAILGRR228 pKa = 11.84SYY230 pKa = 11.41LVGRR234 pKa = 11.84NN235 pKa = 3.28
Molecular weight: 26.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
994 |
235 |
417 |
331.3 |
36.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.841 ± 1.425 | 1.911 ± 0.456 |
5.332 ± 0.485 | 4.628 ± 0.74 |
5.131 ± 0.747 | 6.64 ± 0.58 |
1.107 ± 0.254 | 5.634 ± 0.169 |
5.332 ± 0.797 | 11.066 ± 1.581 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.918 ± 0.211 | 4.225 ± 0.821 |
4.93 ± 1.288 | 2.213 ± 0.315 |
6.942 ± 1.39 | 9.557 ± 0.237 |
6.237 ± 1.986 | 5.231 ± 0.774 |
1.207 ± 0.223 | 2.918 ± 0.61 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
