
Lepus americanus faeces associated microvirus SHP1 6472
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.19
Get precalculated fractions of proteins
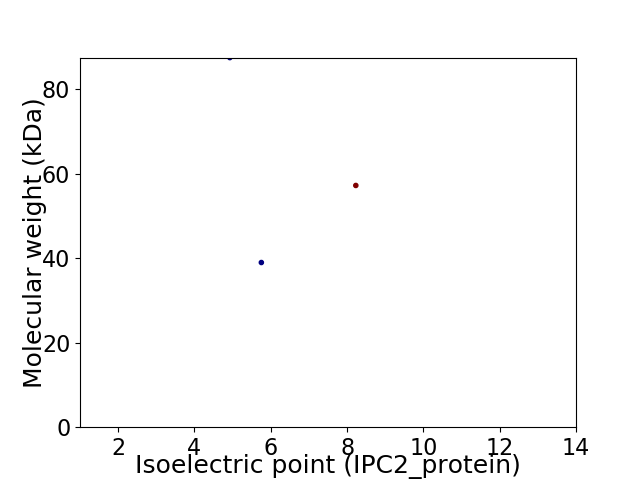
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Z5CJ84|A0A2Z5CJ84_9VIRU Major capsid protein OS=Lepus americanus faeces associated microvirus SHP1 6472 OX=2219217 PE=3 SV=1
MM1 pKa = 7.4SGVFDD6 pKa = 3.7SLKK9 pKa = 10.89NVTNPINRR17 pKa = 11.84NTFDD21 pKa = 3.25WSHH24 pKa = 5.85VNNFTTGFGRR34 pKa = 11.84LTPVLCEE41 pKa = 3.75YY42 pKa = 10.6CPPNTSLRR50 pKa = 11.84IKK52 pKa = 9.76PQFGLRR58 pKa = 11.84FMPMMFPVQTRR69 pKa = 11.84IKK71 pKa = 10.12AYY73 pKa = 9.8LSFYY77 pKa = 10.01RR78 pKa = 11.84VPIRR82 pKa = 11.84TLWKK86 pKa = 10.24DD87 pKa = 3.3YY88 pKa = 10.96KK89 pKa = 11.03DD90 pKa = 3.65WVSSANDD97 pKa = 3.3QTSDD101 pKa = 4.5LEE103 pKa = 4.29PPYY106 pKa = 10.82LDD108 pKa = 5.44LPIDD112 pKa = 4.35SYY114 pKa = 11.36TSGYY118 pKa = 8.36MRR120 pKa = 11.84PSGLADD126 pKa = 3.38YY127 pKa = 11.03LGIPNDD133 pKa = 3.14ITFNDD138 pKa = 3.88SDD140 pKa = 4.11LSSWTISTVSQLNTSNTSSLVNTKK164 pKa = 10.24DD165 pKa = 3.32YY166 pKa = 11.55YY167 pKa = 11.28FFGVSNLMNPAINTGNGIFSYY188 pKa = 10.31EE189 pKa = 3.8LNYY192 pKa = 9.89QRR194 pKa = 11.84NISGYY199 pKa = 8.04PNSAYY204 pKa = 10.84VLATFSQPTFNFRR217 pKa = 11.84GTLKK221 pKa = 10.65FSFSTTQSSAEE232 pKa = 4.02AIINDD237 pKa = 3.49VKK239 pKa = 11.13KK240 pKa = 10.63NKK242 pKa = 8.28TWSICLSTFGQQAAQYY258 pKa = 10.29YY259 pKa = 9.91NYY261 pKa = 10.59TNINPILTVSSSMLSIEE278 pKa = 4.43NNTDD282 pKa = 2.97SDD284 pKa = 4.18STSKK288 pKa = 10.52PKK290 pKa = 10.59KK291 pKa = 9.97LVLTIPSFSFSVSSVPTNSGIVFQYY316 pKa = 10.42RR317 pKa = 11.84SLMLFWRR324 pKa = 11.84DD325 pKa = 2.92HH326 pKa = 6.65SYY328 pKa = 11.35NGSNNDD334 pKa = 3.18EE335 pKa = 4.1NSIKK339 pKa = 10.3EE340 pKa = 3.8FSITQSTNSNLSNQPYY356 pKa = 9.74YY357 pKa = 11.06SSDD360 pKa = 3.18LSAPDD365 pKa = 3.12AKK367 pKa = 11.06YY368 pKa = 10.47GIKK371 pKa = 10.43LSAYY375 pKa = 8.31PFRR378 pKa = 11.84AYY380 pKa = 9.9EE381 pKa = 3.95AVYY384 pKa = 10.26NAYY387 pKa = 9.4IRR389 pKa = 11.84NNKK392 pKa = 8.75NNPFYY397 pKa = 10.59IDD399 pKa = 4.86DD400 pKa = 3.79KK401 pKa = 11.06VSYY404 pKa = 10.35NKK406 pKa = 9.86WIPNDD411 pKa = 3.22NGGADD416 pKa = 3.03KK417 pKa = 10.95YY418 pKa = 11.09AYY420 pKa = 9.47QLHH423 pKa = 6.3HH424 pKa = 7.12SNWASDD430 pKa = 3.66QFTTAVPSPQQGQAPLVGITTYY452 pKa = 10.14TKK454 pKa = 10.21TVALEE459 pKa = 4.25NGHH462 pKa = 6.24SEE464 pKa = 4.43TTVGTALVDD473 pKa = 3.7EE474 pKa = 5.74DD475 pKa = 4.07GNKK478 pKa = 10.19YY479 pKa = 10.3DD480 pKa = 4.91VSYY483 pKa = 10.86EE484 pKa = 4.3SNGDD488 pKa = 3.38EE489 pKa = 4.69LKK491 pKa = 10.78DD492 pKa = 3.82VKK494 pKa = 10.31YY495 pKa = 9.14TLLNADD501 pKa = 3.62TPVRR505 pKa = 11.84PVSNLFDD512 pKa = 4.26LVTSGISINDD522 pKa = 3.28FRR524 pKa = 11.84NVNAYY529 pKa = 8.79QRR531 pKa = 11.84YY532 pKa = 9.51LEE534 pKa = 4.28LNQMRR539 pKa = 11.84GYY541 pKa = 9.64SYY543 pKa = 11.36KK544 pKa = 10.81DD545 pKa = 2.93IVEE548 pKa = 4.05GRR550 pKa = 11.84FDD552 pKa = 3.79VNVRR556 pKa = 11.84YY557 pKa = 10.13DD558 pKa = 3.95DD559 pKa = 5.05LNMPEE564 pKa = 4.32YY565 pKa = 10.43LGGMTRR571 pKa = 11.84DD572 pKa = 3.61VSISPVTQTQAITPGSWQEE591 pKa = 4.07GTYY594 pKa = 8.88STQLGAQAGNGTLFADD610 pKa = 5.06GEE612 pKa = 4.55NISCFCDD619 pKa = 3.1EE620 pKa = 4.33EE621 pKa = 5.43SFVIGILSVCPMPVYY636 pKa = 9.78TQVLPKK642 pKa = 10.4HH643 pKa = 5.64FLYY646 pKa = 10.35RR647 pKa = 11.84DD648 pKa = 3.44RR649 pKa = 11.84LDD651 pKa = 3.52YY652 pKa = 9.95FAPEE656 pKa = 3.75FDD658 pKa = 4.86HH659 pKa = 8.47IGFQPIPLKK668 pKa = 10.62EE669 pKa = 3.7IAPINTYY676 pKa = 8.36FTLGRR681 pKa = 11.84NHH683 pKa = 7.3LDD685 pKa = 3.18DD686 pKa = 3.57TFGYY690 pKa = 8.8QRR692 pKa = 11.84PWYY695 pKa = 8.6EE696 pKa = 3.89FCQKK700 pKa = 10.69LDD702 pKa = 3.52TAHH705 pKa = 7.12GLFKK709 pKa = 10.61TDD711 pKa = 4.46LRR713 pKa = 11.84NFLINRR719 pKa = 11.84VFGGVPSLNSAFTTMDD735 pKa = 3.58EE736 pKa = 4.19NDD738 pKa = 4.09LNDD741 pKa = 3.46VFAVTDD747 pKa = 3.59ISDD750 pKa = 4.51KK751 pKa = 10.88ILGAIWFDD759 pKa = 3.28ITAKK763 pKa = 10.72LPISRR768 pKa = 11.84VSVPRR773 pKa = 11.84LEE775 pKa = 4.1
MM1 pKa = 7.4SGVFDD6 pKa = 3.7SLKK9 pKa = 10.89NVTNPINRR17 pKa = 11.84NTFDD21 pKa = 3.25WSHH24 pKa = 5.85VNNFTTGFGRR34 pKa = 11.84LTPVLCEE41 pKa = 3.75YY42 pKa = 10.6CPPNTSLRR50 pKa = 11.84IKK52 pKa = 9.76PQFGLRR58 pKa = 11.84FMPMMFPVQTRR69 pKa = 11.84IKK71 pKa = 10.12AYY73 pKa = 9.8LSFYY77 pKa = 10.01RR78 pKa = 11.84VPIRR82 pKa = 11.84TLWKK86 pKa = 10.24DD87 pKa = 3.3YY88 pKa = 10.96KK89 pKa = 11.03DD90 pKa = 3.65WVSSANDD97 pKa = 3.3QTSDD101 pKa = 4.5LEE103 pKa = 4.29PPYY106 pKa = 10.82LDD108 pKa = 5.44LPIDD112 pKa = 4.35SYY114 pKa = 11.36TSGYY118 pKa = 8.36MRR120 pKa = 11.84PSGLADD126 pKa = 3.38YY127 pKa = 11.03LGIPNDD133 pKa = 3.14ITFNDD138 pKa = 3.88SDD140 pKa = 4.11LSSWTISTVSQLNTSNTSSLVNTKK164 pKa = 10.24DD165 pKa = 3.32YY166 pKa = 11.55YY167 pKa = 11.28FFGVSNLMNPAINTGNGIFSYY188 pKa = 10.31EE189 pKa = 3.8LNYY192 pKa = 9.89QRR194 pKa = 11.84NISGYY199 pKa = 8.04PNSAYY204 pKa = 10.84VLATFSQPTFNFRR217 pKa = 11.84GTLKK221 pKa = 10.65FSFSTTQSSAEE232 pKa = 4.02AIINDD237 pKa = 3.49VKK239 pKa = 11.13KK240 pKa = 10.63NKK242 pKa = 8.28TWSICLSTFGQQAAQYY258 pKa = 10.29YY259 pKa = 9.91NYY261 pKa = 10.59TNINPILTVSSSMLSIEE278 pKa = 4.43NNTDD282 pKa = 2.97SDD284 pKa = 4.18STSKK288 pKa = 10.52PKK290 pKa = 10.59KK291 pKa = 9.97LVLTIPSFSFSVSSVPTNSGIVFQYY316 pKa = 10.42RR317 pKa = 11.84SLMLFWRR324 pKa = 11.84DD325 pKa = 2.92HH326 pKa = 6.65SYY328 pKa = 11.35NGSNNDD334 pKa = 3.18EE335 pKa = 4.1NSIKK339 pKa = 10.3EE340 pKa = 3.8FSITQSTNSNLSNQPYY356 pKa = 9.74YY357 pKa = 11.06SSDD360 pKa = 3.18LSAPDD365 pKa = 3.12AKK367 pKa = 11.06YY368 pKa = 10.47GIKK371 pKa = 10.43LSAYY375 pKa = 8.31PFRR378 pKa = 11.84AYY380 pKa = 9.9EE381 pKa = 3.95AVYY384 pKa = 10.26NAYY387 pKa = 9.4IRR389 pKa = 11.84NNKK392 pKa = 8.75NNPFYY397 pKa = 10.59IDD399 pKa = 4.86DD400 pKa = 3.79KK401 pKa = 11.06VSYY404 pKa = 10.35NKK406 pKa = 9.86WIPNDD411 pKa = 3.22NGGADD416 pKa = 3.03KK417 pKa = 10.95YY418 pKa = 11.09AYY420 pKa = 9.47QLHH423 pKa = 6.3HH424 pKa = 7.12SNWASDD430 pKa = 3.66QFTTAVPSPQQGQAPLVGITTYY452 pKa = 10.14TKK454 pKa = 10.21TVALEE459 pKa = 4.25NGHH462 pKa = 6.24SEE464 pKa = 4.43TTVGTALVDD473 pKa = 3.7EE474 pKa = 5.74DD475 pKa = 4.07GNKK478 pKa = 10.19YY479 pKa = 10.3DD480 pKa = 4.91VSYY483 pKa = 10.86EE484 pKa = 4.3SNGDD488 pKa = 3.38EE489 pKa = 4.69LKK491 pKa = 10.78DD492 pKa = 3.82VKK494 pKa = 10.31YY495 pKa = 9.14TLLNADD501 pKa = 3.62TPVRR505 pKa = 11.84PVSNLFDD512 pKa = 4.26LVTSGISINDD522 pKa = 3.28FRR524 pKa = 11.84NVNAYY529 pKa = 8.79QRR531 pKa = 11.84YY532 pKa = 9.51LEE534 pKa = 4.28LNQMRR539 pKa = 11.84GYY541 pKa = 9.64SYY543 pKa = 11.36KK544 pKa = 10.81DD545 pKa = 2.93IVEE548 pKa = 4.05GRR550 pKa = 11.84FDD552 pKa = 3.79VNVRR556 pKa = 11.84YY557 pKa = 10.13DD558 pKa = 3.95DD559 pKa = 5.05LNMPEE564 pKa = 4.32YY565 pKa = 10.43LGGMTRR571 pKa = 11.84DD572 pKa = 3.61VSISPVTQTQAITPGSWQEE591 pKa = 4.07GTYY594 pKa = 8.88STQLGAQAGNGTLFADD610 pKa = 5.06GEE612 pKa = 4.55NISCFCDD619 pKa = 3.1EE620 pKa = 4.33EE621 pKa = 5.43SFVIGILSVCPMPVYY636 pKa = 9.78TQVLPKK642 pKa = 10.4HH643 pKa = 5.64FLYY646 pKa = 10.35RR647 pKa = 11.84DD648 pKa = 3.44RR649 pKa = 11.84LDD651 pKa = 3.52YY652 pKa = 9.95FAPEE656 pKa = 3.75FDD658 pKa = 4.86HH659 pKa = 8.47IGFQPIPLKK668 pKa = 10.62EE669 pKa = 3.7IAPINTYY676 pKa = 8.36FTLGRR681 pKa = 11.84NHH683 pKa = 7.3LDD685 pKa = 3.18DD686 pKa = 3.57TFGYY690 pKa = 8.8QRR692 pKa = 11.84PWYY695 pKa = 8.6EE696 pKa = 3.89FCQKK700 pKa = 10.69LDD702 pKa = 3.52TAHH705 pKa = 7.12GLFKK709 pKa = 10.61TDD711 pKa = 4.46LRR713 pKa = 11.84NFLINRR719 pKa = 11.84VFGGVPSLNSAFTTMDD735 pKa = 3.58EE736 pKa = 4.19NDD738 pKa = 4.09LNDD741 pKa = 3.46VFAVTDD747 pKa = 3.59ISDD750 pKa = 4.51KK751 pKa = 10.88ILGAIWFDD759 pKa = 3.28ITAKK763 pKa = 10.72LPISRR768 pKa = 11.84VSVPRR773 pKa = 11.84LEE775 pKa = 4.1
Molecular weight: 87.46 kDa
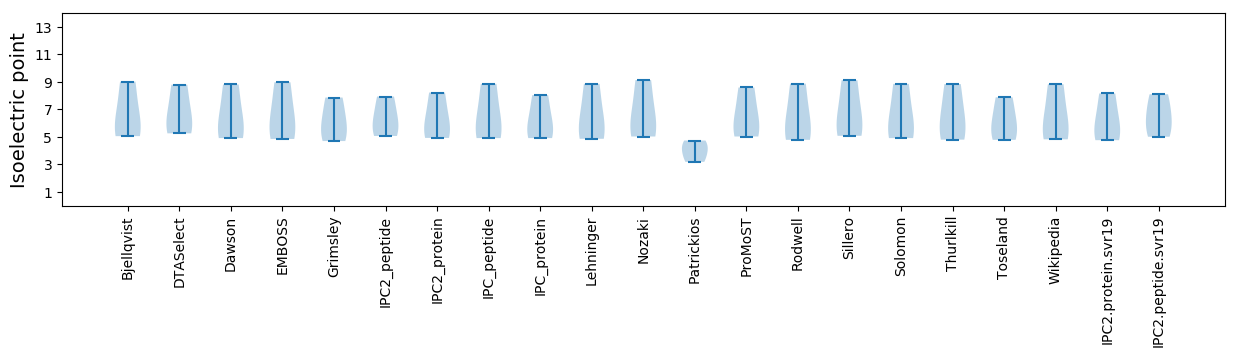
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z5CJ84|A0A2Z5CJ84_9VIRU Major capsid protein OS=Lepus americanus faeces associated microvirus SHP1 6472 OX=2219217 PE=3 SV=1
MM1 pKa = 7.13FCEE4 pKa = 4.23NPVILLNPKK13 pKa = 9.11LRR15 pKa = 11.84KK16 pKa = 8.61LTVHH20 pKa = 7.1CDD22 pKa = 3.55YY23 pKa = 10.44MHH25 pKa = 7.03SPLGDD30 pKa = 3.29YY31 pKa = 11.09SLLDD35 pKa = 3.17KK36 pKa = 10.98RR37 pKa = 11.84LFDD40 pKa = 4.11FRR42 pKa = 11.84RR43 pKa = 11.84VRR45 pKa = 11.84PKK47 pKa = 10.29ICDD50 pKa = 3.36KK51 pKa = 10.13LTKK54 pKa = 10.3DD55 pKa = 3.29NYY57 pKa = 10.9NEE59 pKa = 3.9YY60 pKa = 10.36FVANSSTGEE69 pKa = 4.11TFPLYY74 pKa = 10.39ILVPCGKK81 pKa = 10.23CPLCRR86 pKa = 11.84LTKK89 pKa = 10.36KK90 pKa = 10.64NEE92 pKa = 3.67LAFRR96 pKa = 11.84CVAEE100 pKa = 4.29TNRR103 pKa = 11.84FDD105 pKa = 3.94TLPLFITLTYY115 pKa = 11.04NNAALPDD122 pKa = 4.11CGVCKK127 pKa = 10.57ADD129 pKa = 3.2VQRR132 pKa = 11.84FLKK135 pKa = 10.41RR136 pKa = 11.84LRR138 pKa = 11.84ARR140 pKa = 11.84LDD142 pKa = 3.41YY143 pKa = 11.16EE144 pKa = 4.48KK145 pKa = 10.33IEE147 pKa = 4.17HH148 pKa = 6.14NLRR151 pKa = 11.84YY152 pKa = 9.99LCVSEE157 pKa = 4.15YY158 pKa = 10.66GKK160 pKa = 10.84SFGRR164 pKa = 11.84PHH166 pKa = 7.2YY167 pKa = 10.95HH168 pKa = 5.19MLLWNFPVSAFPNISAVVHH187 pKa = 6.18FIEE190 pKa = 5.12RR191 pKa = 11.84AWSVYY196 pKa = 10.09LLSVPCSKK204 pKa = 10.4RR205 pKa = 11.84VSLSRR210 pKa = 11.84AGEE213 pKa = 3.99LFRR216 pKa = 11.84FTSGRR221 pKa = 11.84RR222 pKa = 11.84QFDD225 pKa = 3.47AVSLGFVYY233 pKa = 10.13VLPLKK238 pKa = 10.55EE239 pKa = 4.28GGVDD243 pKa = 3.63YY244 pKa = 10.95VLKK247 pKa = 11.29YY248 pKa = 10.04MMKK251 pKa = 10.63DD252 pKa = 2.99KK253 pKa = 11.21DD254 pKa = 3.84AVPCGKK260 pKa = 9.85NPLFMLSSRR269 pKa = 11.84KK270 pKa = 9.81NGGIGSEE277 pKa = 4.4YY278 pKa = 10.04IRR280 pKa = 11.84SQRR283 pKa = 11.84DD284 pKa = 3.22YY285 pKa = 11.06FLSAKK290 pKa = 10.48NITTLSIVDD299 pKa = 3.76KK300 pKa = 10.18LVSGRR305 pKa = 11.84VVKK308 pKa = 9.62TKK310 pKa = 8.4ITPYY314 pKa = 10.02IKK316 pKa = 10.26SLLMPSPSRR325 pKa = 11.84KK326 pKa = 9.08YY327 pKa = 10.01RR328 pKa = 11.84RR329 pKa = 11.84KK330 pKa = 9.86EE331 pKa = 3.61YY332 pKa = 11.07NLMKK336 pKa = 10.12EE337 pKa = 4.97FCQKK341 pKa = 10.45LSKK344 pKa = 10.54FMNLCVKK351 pKa = 10.39FNEE354 pKa = 4.42LYY356 pKa = 10.75KK357 pKa = 10.53PIGYY361 pKa = 9.42SRR363 pKa = 11.84EE364 pKa = 4.18TMDD367 pKa = 4.18EE368 pKa = 4.18YY369 pKa = 11.67YY370 pKa = 10.83EE371 pKa = 4.55PFTDD375 pKa = 3.89IYY377 pKa = 11.15SRR379 pKa = 11.84PIWTEE384 pKa = 3.4TLRR387 pKa = 11.84KK388 pKa = 9.49CRR390 pKa = 11.84FLGYY394 pKa = 9.81VPEE397 pKa = 4.93RR398 pKa = 11.84EE399 pKa = 4.04VDD401 pKa = 5.03DD402 pKa = 4.87DD403 pKa = 4.55YY404 pKa = 12.32SWIKK408 pKa = 11.11DD409 pKa = 3.16KK410 pKa = 10.72DD411 pKa = 3.32TWNQFAYY418 pKa = 10.06EE419 pKa = 4.29CLDD422 pKa = 3.52EE423 pKa = 6.05LEE425 pKa = 4.33NLCIKK430 pKa = 10.47ILDD433 pKa = 3.84IPDD436 pKa = 3.05YY437 pKa = 10.8NAYY440 pKa = 10.22FDD442 pKa = 5.56ARR444 pKa = 11.84EE445 pKa = 3.75QYY447 pKa = 11.04LNEE450 pKa = 4.65RR451 pKa = 11.84GLQLEE456 pKa = 4.24YY457 pKa = 9.94EE458 pKa = 4.61YY459 pKa = 11.6CNMTPADD466 pKa = 3.92PHH468 pKa = 7.13VLADD472 pKa = 4.27KK473 pKa = 10.43IRR475 pKa = 11.84YY476 pKa = 8.97SMARR480 pKa = 11.84ALTKK484 pKa = 10.6EE485 pKa = 4.29KK486 pKa = 10.51II487 pKa = 3.62
MM1 pKa = 7.13FCEE4 pKa = 4.23NPVILLNPKK13 pKa = 9.11LRR15 pKa = 11.84KK16 pKa = 8.61LTVHH20 pKa = 7.1CDD22 pKa = 3.55YY23 pKa = 10.44MHH25 pKa = 7.03SPLGDD30 pKa = 3.29YY31 pKa = 11.09SLLDD35 pKa = 3.17KK36 pKa = 10.98RR37 pKa = 11.84LFDD40 pKa = 4.11FRR42 pKa = 11.84RR43 pKa = 11.84VRR45 pKa = 11.84PKK47 pKa = 10.29ICDD50 pKa = 3.36KK51 pKa = 10.13LTKK54 pKa = 10.3DD55 pKa = 3.29NYY57 pKa = 10.9NEE59 pKa = 3.9YY60 pKa = 10.36FVANSSTGEE69 pKa = 4.11TFPLYY74 pKa = 10.39ILVPCGKK81 pKa = 10.23CPLCRR86 pKa = 11.84LTKK89 pKa = 10.36KK90 pKa = 10.64NEE92 pKa = 3.67LAFRR96 pKa = 11.84CVAEE100 pKa = 4.29TNRR103 pKa = 11.84FDD105 pKa = 3.94TLPLFITLTYY115 pKa = 11.04NNAALPDD122 pKa = 4.11CGVCKK127 pKa = 10.57ADD129 pKa = 3.2VQRR132 pKa = 11.84FLKK135 pKa = 10.41RR136 pKa = 11.84LRR138 pKa = 11.84ARR140 pKa = 11.84LDD142 pKa = 3.41YY143 pKa = 11.16EE144 pKa = 4.48KK145 pKa = 10.33IEE147 pKa = 4.17HH148 pKa = 6.14NLRR151 pKa = 11.84YY152 pKa = 9.99LCVSEE157 pKa = 4.15YY158 pKa = 10.66GKK160 pKa = 10.84SFGRR164 pKa = 11.84PHH166 pKa = 7.2YY167 pKa = 10.95HH168 pKa = 5.19MLLWNFPVSAFPNISAVVHH187 pKa = 6.18FIEE190 pKa = 5.12RR191 pKa = 11.84AWSVYY196 pKa = 10.09LLSVPCSKK204 pKa = 10.4RR205 pKa = 11.84VSLSRR210 pKa = 11.84AGEE213 pKa = 3.99LFRR216 pKa = 11.84FTSGRR221 pKa = 11.84RR222 pKa = 11.84QFDD225 pKa = 3.47AVSLGFVYY233 pKa = 10.13VLPLKK238 pKa = 10.55EE239 pKa = 4.28GGVDD243 pKa = 3.63YY244 pKa = 10.95VLKK247 pKa = 11.29YY248 pKa = 10.04MMKK251 pKa = 10.63DD252 pKa = 2.99KK253 pKa = 11.21DD254 pKa = 3.84AVPCGKK260 pKa = 9.85NPLFMLSSRR269 pKa = 11.84KK270 pKa = 9.81NGGIGSEE277 pKa = 4.4YY278 pKa = 10.04IRR280 pKa = 11.84SQRR283 pKa = 11.84DD284 pKa = 3.22YY285 pKa = 11.06FLSAKK290 pKa = 10.48NITTLSIVDD299 pKa = 3.76KK300 pKa = 10.18LVSGRR305 pKa = 11.84VVKK308 pKa = 9.62TKK310 pKa = 8.4ITPYY314 pKa = 10.02IKK316 pKa = 10.26SLLMPSPSRR325 pKa = 11.84KK326 pKa = 9.08YY327 pKa = 10.01RR328 pKa = 11.84RR329 pKa = 11.84KK330 pKa = 9.86EE331 pKa = 3.61YY332 pKa = 11.07NLMKK336 pKa = 10.12EE337 pKa = 4.97FCQKK341 pKa = 10.45LSKK344 pKa = 10.54FMNLCVKK351 pKa = 10.39FNEE354 pKa = 4.42LYY356 pKa = 10.75KK357 pKa = 10.53PIGYY361 pKa = 9.42SRR363 pKa = 11.84EE364 pKa = 4.18TMDD367 pKa = 4.18EE368 pKa = 4.18YY369 pKa = 11.67YY370 pKa = 10.83EE371 pKa = 4.55PFTDD375 pKa = 3.89IYY377 pKa = 11.15SRR379 pKa = 11.84PIWTEE384 pKa = 3.4TLRR387 pKa = 11.84KK388 pKa = 9.49CRR390 pKa = 11.84FLGYY394 pKa = 9.81VPEE397 pKa = 4.93RR398 pKa = 11.84EE399 pKa = 4.04VDD401 pKa = 5.03DD402 pKa = 4.87DD403 pKa = 4.55YY404 pKa = 12.32SWIKK408 pKa = 11.11DD409 pKa = 3.16KK410 pKa = 10.72DD411 pKa = 3.32TWNQFAYY418 pKa = 10.06EE419 pKa = 4.29CLDD422 pKa = 3.52EE423 pKa = 6.05LEE425 pKa = 4.33NLCIKK430 pKa = 10.47ILDD433 pKa = 3.84IPDD436 pKa = 3.05YY437 pKa = 10.8NAYY440 pKa = 10.22FDD442 pKa = 5.56ARR444 pKa = 11.84EE445 pKa = 3.75QYY447 pKa = 11.04LNEE450 pKa = 4.65RR451 pKa = 11.84GLQLEE456 pKa = 4.24YY457 pKa = 9.94EE458 pKa = 4.61YY459 pKa = 11.6CNMTPADD466 pKa = 3.92PHH468 pKa = 7.13VLADD472 pKa = 4.27KK473 pKa = 10.43IRR475 pKa = 11.84YY476 pKa = 8.97SMARR480 pKa = 11.84ALTKK484 pKa = 10.6EE485 pKa = 4.29KK486 pKa = 10.51II487 pKa = 3.62
Molecular weight: 57.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1626 |
364 |
775 |
542.0 |
61.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.027 ± 1.663 | 1.722 ± 0.714 |
6.335 ± 0.319 | 4.428 ± 0.754 |
4.859 ± 0.887 | 5.228 ± 0.488 |
1.107 ± 0.178 | 5.72 ± 0.562 |
5.781 ± 1.115 | 9.41 ± 1.035 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.03 ± 0.22 | 7.196 ± 0.921 |
4.613 ± 1.061 | 3.813 ± 1.105 |
4.797 ± 0.936 | 9.287 ± 1.147 |
5.904 ± 1.237 | 5.535 ± 0.359 |
1.046 ± 0.286 | 5.166 ± 1.278 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
