
Streptococcus phage Javan273
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.48
Get precalculated fractions of proteins
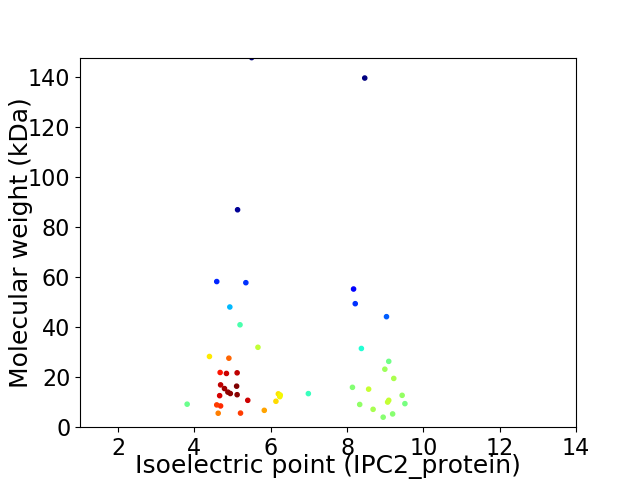
Virtual 2D-PAGE plot for 49 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
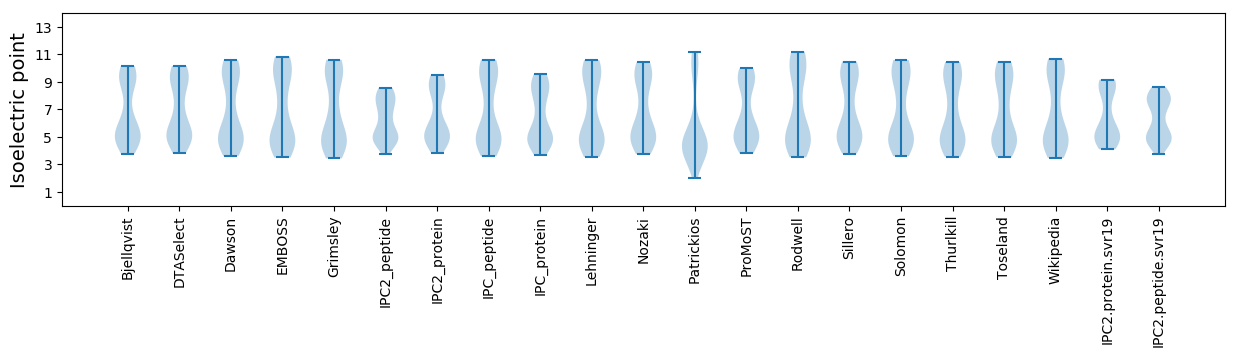
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4D6A7F1|A0A4D6A7F1_9CAUD Tail protein OS=Streptococcus phage Javan273 OX=2548088 GN=Javan273_0014 PE=4 SV=1
MM1 pKa = 6.92ATKK4 pKa = 10.54EE5 pKa = 3.9EE6 pKa = 4.35VIAFAKK12 pKa = 10.5NLADD16 pKa = 3.48TGQGVDD22 pKa = 4.87LDD24 pKa = 4.72GVWGTQCVDD33 pKa = 3.53LPNWITTKK41 pKa = 10.41YY42 pKa = 10.5FSVALWGNAIDD53 pKa = 5.36LLDD56 pKa = 3.79SAKK59 pKa = 10.31AQGMEE64 pKa = 4.17VVYY67 pKa = 10.06DD68 pKa = 3.81APGVNPRR75 pKa = 11.84DD76 pKa = 3.23GAIFVMVTYY85 pKa = 10.75AHH87 pKa = 7.21GYY89 pKa = 7.86GHH91 pKa = 7.22TGLVIQTSDD100 pKa = 3.4GYY102 pKa = 11.11VLYY105 pKa = 10.01TIEE108 pKa = 4.29QNVDD112 pKa = 3.0GYY114 pKa = 11.54SDD116 pKa = 3.84NNGDD120 pKa = 4.5GINDD124 pKa = 3.41QLQFGGPARR133 pKa = 11.84YY134 pKa = 9.36INRR137 pKa = 11.84PFSDD141 pKa = 3.17GTGYY145 pKa = 10.47ILGWFYY151 pKa = 10.79PPYY154 pKa = 10.67NNTPAQQLEE163 pKa = 4.23TSAPVKK169 pKa = 10.64SEE171 pKa = 3.8SDD173 pKa = 3.16GTYY176 pKa = 9.8VANPEE181 pKa = 4.2TGTFTVRR188 pKa = 11.84VAALNVRR195 pKa = 11.84SAPRR199 pKa = 11.84LDD201 pKa = 3.6AEE203 pKa = 4.28IVATYY208 pKa = 9.87GEE210 pKa = 4.27NMEE213 pKa = 4.47FNYY216 pKa = 10.8DD217 pKa = 3.03GWLDD221 pKa = 3.37SDD223 pKa = 4.15GYY225 pKa = 10.74IWVTYY230 pKa = 10.42ISATGIRR237 pKa = 11.84RR238 pKa = 11.84YY239 pKa = 10.09VAVGNSKK246 pKa = 10.33NGRR249 pKa = 11.84RR250 pKa = 11.84VTNFGTFRR258 pKa = 4.37
MM1 pKa = 6.92ATKK4 pKa = 10.54EE5 pKa = 3.9EE6 pKa = 4.35VIAFAKK12 pKa = 10.5NLADD16 pKa = 3.48TGQGVDD22 pKa = 4.87LDD24 pKa = 4.72GVWGTQCVDD33 pKa = 3.53LPNWITTKK41 pKa = 10.41YY42 pKa = 10.5FSVALWGNAIDD53 pKa = 5.36LLDD56 pKa = 3.79SAKK59 pKa = 10.31AQGMEE64 pKa = 4.17VVYY67 pKa = 10.06DD68 pKa = 3.81APGVNPRR75 pKa = 11.84DD76 pKa = 3.23GAIFVMVTYY85 pKa = 10.75AHH87 pKa = 7.21GYY89 pKa = 7.86GHH91 pKa = 7.22TGLVIQTSDD100 pKa = 3.4GYY102 pKa = 11.11VLYY105 pKa = 10.01TIEE108 pKa = 4.29QNVDD112 pKa = 3.0GYY114 pKa = 11.54SDD116 pKa = 3.84NNGDD120 pKa = 4.5GINDD124 pKa = 3.41QLQFGGPARR133 pKa = 11.84YY134 pKa = 9.36INRR137 pKa = 11.84PFSDD141 pKa = 3.17GTGYY145 pKa = 10.47ILGWFYY151 pKa = 10.79PPYY154 pKa = 10.67NNTPAQQLEE163 pKa = 4.23TSAPVKK169 pKa = 10.64SEE171 pKa = 3.8SDD173 pKa = 3.16GTYY176 pKa = 9.8VANPEE181 pKa = 4.2TGTFTVRR188 pKa = 11.84VAALNVRR195 pKa = 11.84SAPRR199 pKa = 11.84LDD201 pKa = 3.6AEE203 pKa = 4.28IVATYY208 pKa = 9.87GEE210 pKa = 4.27NMEE213 pKa = 4.47FNYY216 pKa = 10.8DD217 pKa = 3.03GWLDD221 pKa = 3.37SDD223 pKa = 4.15GYY225 pKa = 10.74IWVTYY230 pKa = 10.42ISATGIRR237 pKa = 11.84RR238 pKa = 11.84YY239 pKa = 10.09VAVGNSKK246 pKa = 10.33NGRR249 pKa = 11.84RR250 pKa = 11.84VTNFGTFRR258 pKa = 4.37
Molecular weight: 28.25 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4D6AB52|A0A4D6AB52_9CAUD Terminase small subunit OS=Streptococcus phage Javan273 OX=2548088 GN=Javan273_0026 PE=4 SV=1
MM1 pKa = 7.84RR2 pKa = 11.84GLDD5 pKa = 3.68EE6 pKa = 5.22FIKK9 pKa = 10.62HH10 pKa = 6.16AMKK13 pKa = 10.43QKK15 pKa = 10.51QEE17 pKa = 3.98VKK19 pKa = 10.25SAVDD23 pKa = 3.71FEE25 pKa = 4.66IGQAALRR32 pKa = 11.84VEE34 pKa = 4.33RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09ILAPVDD44 pKa = 3.77TGWLRR49 pKa = 11.84TQIYY53 pKa = 10.01NEE55 pKa = 4.27KK56 pKa = 9.58QRR58 pKa = 11.84MMHH61 pKa = 5.91YY62 pKa = 10.3KK63 pKa = 10.15VISPALYY70 pKa = 9.93SVYY73 pKa = 10.79LEE75 pKa = 4.19LGTRR79 pKa = 11.84YY80 pKa = 9.16MSAQPYY86 pKa = 10.18LDD88 pKa = 3.74PALRR92 pKa = 11.84AEE94 pKa = 4.57WPVLMANLKK103 pKa = 10.68KK104 pKa = 10.07MFRR107 pKa = 11.84RR108 pKa = 3.88
MM1 pKa = 7.84RR2 pKa = 11.84GLDD5 pKa = 3.68EE6 pKa = 5.22FIKK9 pKa = 10.62HH10 pKa = 6.16AMKK13 pKa = 10.43QKK15 pKa = 10.51QEE17 pKa = 3.98VKK19 pKa = 10.25SAVDD23 pKa = 3.71FEE25 pKa = 4.66IGQAALRR32 pKa = 11.84VEE34 pKa = 4.33RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09ILAPVDD44 pKa = 3.77TGWLRR49 pKa = 11.84TQIYY53 pKa = 10.01NEE55 pKa = 4.27KK56 pKa = 9.58QRR58 pKa = 11.84MMHH61 pKa = 5.91YY62 pKa = 10.3KK63 pKa = 10.15VISPALYY70 pKa = 9.93SVYY73 pKa = 10.79LEE75 pKa = 4.19LGTRR79 pKa = 11.84YY80 pKa = 9.16MSAQPYY86 pKa = 10.18LDD88 pKa = 3.74PALRR92 pKa = 11.84AEE94 pKa = 4.57WPVLMANLKK103 pKa = 10.68KK104 pKa = 10.07MFRR107 pKa = 11.84RR108 pKa = 3.88
Molecular weight: 12.73 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
11584 |
37 |
1364 |
236.4 |
26.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.674 ± 0.728 | 0.423 ± 0.11 |
6.259 ± 0.498 | 6.967 ± 0.591 |
3.626 ± 0.172 | 6.215 ± 0.459 |
1.148 ± 0.163 | 7.096 ± 0.341 |
8.106 ± 0.526 | 7.907 ± 0.299 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.616 ± 0.274 | 6.06 ± 0.335 |
2.823 ± 0.258 | 4.273 ± 0.197 |
4.204 ± 0.342 | 6.854 ± 0.85 |
6.673 ± 0.357 | 6.259 ± 0.249 |
1.183 ± 0.177 | 3.634 ± 0.385 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
