
Thermobaculum terrenum (strain ATCC BAA-798 / YNP1)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Chloroflexi; Chloroflexi incertae sedis; Thermobaculum; Thermobaculum terrenum
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins
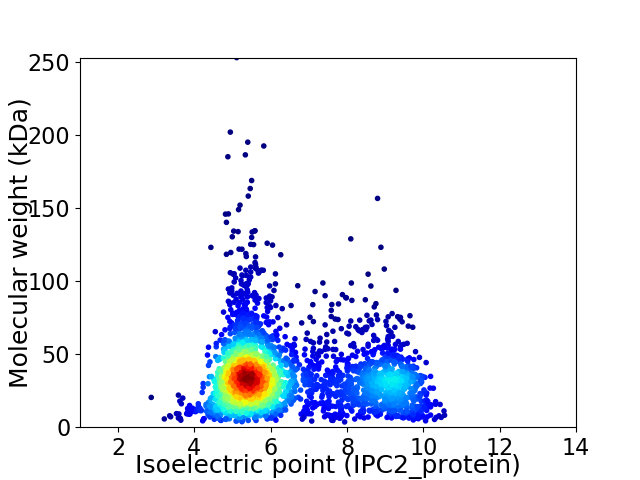
Virtual 2D-PAGE plot for 2827 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D1CGV8|D1CGV8_THET1 Oligopeptide/dipeptide ABC transporter ATPase subunit OS=Thermobaculum terrenum (strain ATCC BAA-798 / YNP1) OX=525904 GN=Tter_2076 PE=4 SV=1
MM1 pKa = 6.98TRR3 pKa = 11.84KK4 pKa = 8.58LTGRR8 pKa = 11.84LATLLMLLAALSMAALATGNSVPPSGLSTSTSSIGPNALKK48 pKa = 10.41PSQCASLNLTNLLVVNGFGFSPHH71 pKa = 6.13VPTLILGSSGPDD83 pKa = 3.25YY84 pKa = 10.96IFAGIDD90 pKa = 3.38DD91 pKa = 4.36DD92 pKa = 5.98CIVAGAGNDD101 pKa = 4.09FISGSDD107 pKa = 3.42GDD109 pKa = 5.13DD110 pKa = 3.66IILAGSGADD119 pKa = 3.64YY120 pKa = 11.51VNGGPGYY127 pKa = 9.65DD128 pKa = 2.69ICYY131 pKa = 10.42GVFDD135 pKa = 5.25PGDD138 pKa = 3.44TAVGCEE144 pKa = 3.82VRR146 pKa = 11.84LPP148 pKa = 3.65
MM1 pKa = 6.98TRR3 pKa = 11.84KK4 pKa = 8.58LTGRR8 pKa = 11.84LATLLMLLAALSMAALATGNSVPPSGLSTSTSSIGPNALKK48 pKa = 10.41PSQCASLNLTNLLVVNGFGFSPHH71 pKa = 6.13VPTLILGSSGPDD83 pKa = 3.25YY84 pKa = 10.96IFAGIDD90 pKa = 3.38DD91 pKa = 4.36DD92 pKa = 5.98CIVAGAGNDD101 pKa = 4.09FISGSDD107 pKa = 3.42GDD109 pKa = 5.13DD110 pKa = 3.66IILAGSGADD119 pKa = 3.64YY120 pKa = 11.51VNGGPGYY127 pKa = 9.65DD128 pKa = 2.69ICYY131 pKa = 10.42GVFDD135 pKa = 5.25PGDD138 pKa = 3.44TAVGCEE144 pKa = 3.82VRR146 pKa = 11.84LPP148 pKa = 3.65
Molecular weight: 14.84 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D1CCJ8|D1CCJ8_THET1 Cell division protein FtsW OS=Thermobaculum terrenum (strain ATCC BAA-798 / YNP1) OX=525904 GN=Tter_1607 PE=4 SV=1
MM1 pKa = 7.38RR2 pKa = 11.84QANGEE7 pKa = 4.03GTVYY11 pKa = 10.44RR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84DD15 pKa = 3.84GRR17 pKa = 11.84WVAAVVHH24 pKa = 6.4AGRR27 pKa = 11.84RR28 pKa = 11.84ISRR31 pKa = 11.84YY32 pKa = 9.09AKK34 pKa = 9.2TRR36 pKa = 11.84RR37 pKa = 11.84EE38 pKa = 3.91AQKK41 pKa = 10.09QLRR44 pKa = 11.84EE45 pKa = 4.08LLEE48 pKa = 4.1AAAEE52 pKa = 4.15HH53 pKa = 6.88RR54 pKa = 11.84LVPPNKK60 pKa = 9.21LTLEE64 pKa = 4.08QYY66 pKa = 9.61LAEE69 pKa = 4.01WLEE72 pKa = 3.91VRR74 pKa = 11.84SRR76 pKa = 11.84EE77 pKa = 4.08LRR79 pKa = 11.84VSTRR83 pKa = 11.84VNYY86 pKa = 9.67QRR88 pKa = 11.84LIEE91 pKa = 4.0QHH93 pKa = 5.95ICPALGGRR101 pKa = 11.84RR102 pKa = 11.84LQALAALEE110 pKa = 4.0LSRR113 pKa = 11.84WLAGLGEE120 pKa = 4.3RR121 pKa = 11.84LPRR124 pKa = 11.84RR125 pKa = 11.84AQEE128 pKa = 4.3AYY130 pKa = 10.54ALLHH134 pKa = 6.46KK135 pKa = 10.66ALADD139 pKa = 3.68AVRR142 pKa = 11.84LGLLATNPLDD152 pKa = 3.61RR153 pKa = 11.84VEE155 pKa = 4.39PPKK158 pKa = 10.21HH159 pKa = 4.65QRR161 pKa = 11.84RR162 pKa = 11.84RR163 pKa = 11.84PTLPARR169 pKa = 11.84DD170 pKa = 3.85QIALLIAALEE180 pKa = 4.18AGKK183 pKa = 10.49AGWYY187 pKa = 10.02SEE189 pKa = 4.82LLLFLLGSGCRR200 pKa = 11.84IGEE203 pKa = 4.02ALGLEE208 pKa = 4.24WGDD211 pKa = 3.62VDD213 pKa = 3.51WQEE216 pKa = 3.78GSVRR220 pKa = 11.84ISRR223 pKa = 11.84QLLEE227 pKa = 4.29VGGEE231 pKa = 4.18VHH233 pKa = 7.04EE234 pKa = 4.91SPPKK238 pKa = 9.38SRR240 pKa = 11.84AGEE243 pKa = 3.92RR244 pKa = 11.84VIALPAFALEE254 pKa = 4.13ALRR257 pKa = 11.84QQRR260 pKa = 11.84AKK262 pKa = 10.93KK263 pKa = 9.55LGKK266 pKa = 10.2YY267 pKa = 9.77CFLSSTGTTPCRR279 pKa = 11.84RR280 pKa = 11.84NVLRR284 pKa = 11.84ALHH287 pKa = 6.19AVCAEE292 pKa = 4.06LGLPRR297 pKa = 11.84LRR299 pKa = 11.84IHH301 pKa = 7.28DD302 pKa = 4.27LRR304 pKa = 11.84HH305 pKa = 4.33VHH307 pKa = 6.99ASLLAHH313 pKa = 6.89AGVPPKK319 pKa = 10.35VAQSRR324 pKa = 11.84LGHH327 pKa = 6.01SSPMVTLQVYY337 pKa = 7.19QHH339 pKa = 6.17VLDD342 pKa = 4.44GADD345 pKa = 3.36RR346 pKa = 11.84EE347 pKa = 4.3AALRR351 pKa = 11.84LEE353 pKa = 4.41RR354 pKa = 11.84VLGG357 pKa = 3.87
MM1 pKa = 7.38RR2 pKa = 11.84QANGEE7 pKa = 4.03GTVYY11 pKa = 10.44RR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84DD15 pKa = 3.84GRR17 pKa = 11.84WVAAVVHH24 pKa = 6.4AGRR27 pKa = 11.84RR28 pKa = 11.84ISRR31 pKa = 11.84YY32 pKa = 9.09AKK34 pKa = 9.2TRR36 pKa = 11.84RR37 pKa = 11.84EE38 pKa = 3.91AQKK41 pKa = 10.09QLRR44 pKa = 11.84EE45 pKa = 4.08LLEE48 pKa = 4.1AAAEE52 pKa = 4.15HH53 pKa = 6.88RR54 pKa = 11.84LVPPNKK60 pKa = 9.21LTLEE64 pKa = 4.08QYY66 pKa = 9.61LAEE69 pKa = 4.01WLEE72 pKa = 3.91VRR74 pKa = 11.84SRR76 pKa = 11.84EE77 pKa = 4.08LRR79 pKa = 11.84VSTRR83 pKa = 11.84VNYY86 pKa = 9.67QRR88 pKa = 11.84LIEE91 pKa = 4.0QHH93 pKa = 5.95ICPALGGRR101 pKa = 11.84RR102 pKa = 11.84LQALAALEE110 pKa = 4.0LSRR113 pKa = 11.84WLAGLGEE120 pKa = 4.3RR121 pKa = 11.84LPRR124 pKa = 11.84RR125 pKa = 11.84AQEE128 pKa = 4.3AYY130 pKa = 10.54ALLHH134 pKa = 6.46KK135 pKa = 10.66ALADD139 pKa = 3.68AVRR142 pKa = 11.84LGLLATNPLDD152 pKa = 3.61RR153 pKa = 11.84VEE155 pKa = 4.39PPKK158 pKa = 10.21HH159 pKa = 4.65QRR161 pKa = 11.84RR162 pKa = 11.84RR163 pKa = 11.84PTLPARR169 pKa = 11.84DD170 pKa = 3.85QIALLIAALEE180 pKa = 4.18AGKK183 pKa = 10.49AGWYY187 pKa = 10.02SEE189 pKa = 4.82LLLFLLGSGCRR200 pKa = 11.84IGEE203 pKa = 4.02ALGLEE208 pKa = 4.24WGDD211 pKa = 3.62VDD213 pKa = 3.51WQEE216 pKa = 3.78GSVRR220 pKa = 11.84ISRR223 pKa = 11.84QLLEE227 pKa = 4.29VGGEE231 pKa = 4.18VHH233 pKa = 7.04EE234 pKa = 4.91SPPKK238 pKa = 9.38SRR240 pKa = 11.84AGEE243 pKa = 3.92RR244 pKa = 11.84VIALPAFALEE254 pKa = 4.13ALRR257 pKa = 11.84QQRR260 pKa = 11.84AKK262 pKa = 10.93KK263 pKa = 9.55LGKK266 pKa = 10.2YY267 pKa = 9.77CFLSSTGTTPCRR279 pKa = 11.84RR280 pKa = 11.84NVLRR284 pKa = 11.84ALHH287 pKa = 6.19AVCAEE292 pKa = 4.06LGLPRR297 pKa = 11.84LRR299 pKa = 11.84IHH301 pKa = 7.28DD302 pKa = 4.27LRR304 pKa = 11.84HH305 pKa = 4.33VHH307 pKa = 6.99ASLLAHH313 pKa = 6.89AGVPPKK319 pKa = 10.35VAQSRR324 pKa = 11.84LGHH327 pKa = 6.01SSPMVTLQVYY337 pKa = 7.19QHH339 pKa = 6.17VLDD342 pKa = 4.44GADD345 pKa = 3.36RR346 pKa = 11.84EE347 pKa = 4.3AALRR351 pKa = 11.84LEE353 pKa = 4.41RR354 pKa = 11.84VLGG357 pKa = 3.87
Molecular weight: 39.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
935529 |
31 |
2240 |
330.9 |
36.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.905 ± 0.044 | 0.891 ± 0.015 |
5.212 ± 0.034 | 6.492 ± 0.054 |
3.321 ± 0.029 | 7.857 ± 0.041 |
2.099 ± 0.02 | 6.107 ± 0.044 |
3.622 ± 0.038 | 10.798 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.291 ± 0.017 | 2.927 ± 0.028 |
5.202 ± 0.035 | 3.484 ± 0.028 |
6.83 ± 0.045 | 6.343 ± 0.049 |
4.963 ± 0.028 | 7.824 ± 0.04 |
1.608 ± 0.022 | 3.223 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
