
Sphingobium sp. RAC03
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Sphingobium; unclassified Sphingobium
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
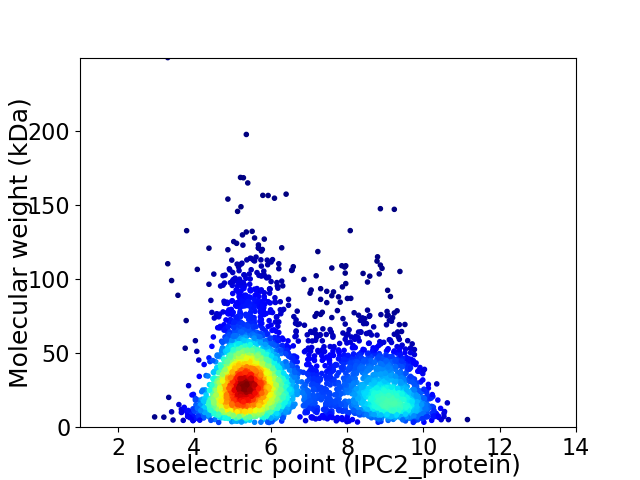
Virtual 2D-PAGE plot for 4109 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
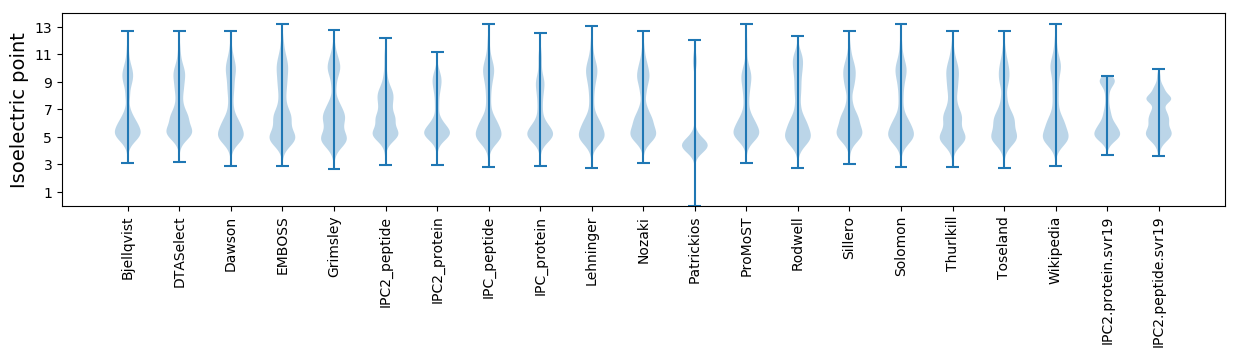
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B3MTR0|A0A1B3MTR0_9SPHN Bacterial regulatory s tetR family protein OS=Sphingobium sp. RAC03 OX=1843368 GN=BSY17_2335 PE=4 SV=1
MM1 pKa = 8.13DD2 pKa = 3.97FTGGAGDD9 pKa = 5.82DD10 pKa = 4.37IYY12 pKa = 11.26TGDD15 pKa = 4.2NSDD18 pKa = 4.14EE19 pKa = 4.42YY20 pKa = 11.01ISGLGGNDD28 pKa = 3.01WLSGGEE34 pKa = 4.11GADD37 pKa = 3.92NIAGGAGADD46 pKa = 3.87RR47 pKa = 11.84LNGGDD52 pKa = 3.54GSDD55 pKa = 3.36VLFSFGRR62 pKa = 11.84TTNMMAPYY70 pKa = 9.6FPLLAGVVSDD80 pKa = 6.4DD81 pKa = 3.47IFADD85 pKa = 3.22VDD87 pKa = 3.97TLIGGAGDD95 pKa = 4.03DD96 pKa = 4.45FFFAGYY102 pKa = 10.04GDD104 pKa = 4.95HH105 pKa = 7.46IDD107 pKa = 3.78GGSYY111 pKa = 11.0DD112 pKa = 3.42SFGNRR117 pKa = 11.84LFISFQGATSGVVADD132 pKa = 6.13FRR134 pKa = 11.84PLQNGEE140 pKa = 4.37SITIGGGIISNIQNIGFLEE159 pKa = 4.11GRR161 pKa = 11.84EE162 pKa = 3.9YY163 pKa = 11.45DD164 pKa = 4.73DD165 pKa = 5.92FIASIDD171 pKa = 3.47TYY173 pKa = 10.32YY174 pKa = 10.57PSGADD179 pKa = 2.77IFGRR183 pKa = 11.84GGNDD187 pKa = 3.4TIIADD192 pKa = 4.39YY193 pKa = 10.58YY194 pKa = 10.4SGWGGAIWGGDD205 pKa = 3.22GDD207 pKa = 4.4DD208 pKa = 4.06TVDD211 pKa = 3.47ATGAQYY217 pKa = 10.78GARR220 pKa = 11.84VYY222 pKa = 11.18GEE224 pKa = 4.46AGNDD228 pKa = 3.82IIRR231 pKa = 11.84MNYY234 pKa = 7.29GTADD238 pKa = 3.59GGDD241 pKa = 3.89GDD243 pKa = 5.19DD244 pKa = 4.32IIYY247 pKa = 10.56GGSAFGGAGNDD258 pKa = 3.33RR259 pKa = 11.84LIDD262 pKa = 3.73VMSGDD267 pKa = 4.21GGDD270 pKa = 3.82GNDD273 pKa = 3.83IITLNFSYY281 pKa = 10.55YY282 pKa = 10.72GNGVATGGRR291 pKa = 11.84GDD293 pKa = 3.86DD294 pKa = 4.71LIRR297 pKa = 11.84GTDD300 pKa = 3.52YY301 pKa = 11.56GSVLVGGEE309 pKa = 4.09GADD312 pKa = 3.63EE313 pKa = 4.41LFGGAGADD321 pKa = 4.17LIYY324 pKa = 10.7TGGRR328 pKa = 11.84NEE330 pKa = 5.18DD331 pKa = 3.35GSALADD337 pKa = 3.36VGVEE341 pKa = 3.85KK342 pKa = 11.08DD343 pKa = 3.98FVSAGAGADD352 pKa = 3.96TIWAGVGDD360 pKa = 4.83DD361 pKa = 3.83VDD363 pKa = 5.12GGDD366 pKa = 4.5GADD369 pKa = 3.06TLYY372 pKa = 11.19YY373 pKa = 10.59SFGGATTGVDD383 pKa = 2.89ISTHH387 pKa = 4.67QFLGATARR395 pKa = 11.84VIGGGTIISIEE406 pKa = 4.09SLAEE410 pKa = 3.75LRR412 pKa = 11.84ASSFDD417 pKa = 3.53DD418 pKa = 4.27RR419 pKa = 11.84IVATTQDD426 pKa = 3.5VRR428 pKa = 11.84LSIYY432 pKa = 10.48GGAGNDD438 pKa = 3.42VVTSSGSSITFRR450 pKa = 11.84GEE452 pKa = 3.59EE453 pKa = 4.09GDD455 pKa = 4.07DD456 pKa = 3.4RR457 pKa = 11.84LISGEE462 pKa = 4.0AADD465 pKa = 4.02MFVGGFGTDD474 pKa = 3.25TIDD477 pKa = 3.4YY478 pKa = 10.37SLYY481 pKa = 8.9TNGVTVTLGLSGAIGIGGGGDD502 pKa = 3.43SLISVEE508 pKa = 4.3NVVGSAFSDD517 pKa = 4.11DD518 pKa = 3.45IQGNEE523 pKa = 4.29LANMLEE529 pKa = 4.46GGAGNDD535 pKa = 4.28TIHH538 pKa = 6.96GGAGDD543 pKa = 4.43DD544 pKa = 3.67RR545 pKa = 11.84LLPGTGHH552 pKa = 6.81NIVDD556 pKa = 4.08GGDD559 pKa = 3.65GFDD562 pKa = 3.6TLVLTGGVASYY573 pKa = 11.23NYY575 pKa = 9.56LVSGASTFVIGEE587 pKa = 4.17EE588 pKa = 4.2GASRR592 pKa = 11.84IVNVEE597 pKa = 3.7HH598 pKa = 6.59VAFANGALGSADD610 pKa = 3.46LAGSLSAFDD619 pKa = 3.73GLRR622 pKa = 11.84YY623 pKa = 8.98IAGYY627 pKa = 10.23GDD629 pKa = 5.42LIAAFGSDD637 pKa = 3.51ADD639 pKa = 3.95SATAHH644 pKa = 5.63YY645 pKa = 9.48VASGFAEE652 pKa = 4.24GRR654 pKa = 11.84DD655 pKa = 3.32AKK657 pKa = 10.91EE658 pKa = 3.92FDD660 pKa = 3.67VLDD663 pKa = 4.37YY664 pKa = 11.01IAGYY668 pKa = 10.5DD669 pKa = 3.71DD670 pKa = 6.12LIAAFGTDD678 pKa = 3.18TQAATSHH685 pKa = 6.3YY686 pKa = 9.32IDD688 pKa = 3.91GGFVEE693 pKa = 5.02GRR695 pKa = 11.84SDD697 pKa = 3.7ALFDD701 pKa = 3.64GLQYY705 pKa = 10.23IASYY709 pKa = 10.92GDD711 pKa = 3.93LIEE714 pKa = 5.77AFGTDD719 pKa = 2.83ADD721 pKa = 3.49AGARR725 pKa = 11.84HH726 pKa = 6.8FIQGGRR732 pKa = 11.84DD733 pKa = 3.2EE734 pKa = 5.16GRR736 pKa = 11.84SDD738 pKa = 4.61DD739 pKa = 5.51LFDD742 pKa = 3.91GLQYY746 pKa = 10.2VASYY750 pKa = 10.81GDD752 pKa = 4.23LIGAIGTDD760 pKa = 3.27VEE762 pKa = 4.28AAAEE766 pKa = 4.05HH767 pKa = 6.67FIKK770 pKa = 10.71NGYY773 pKa = 9.35GEE775 pKa = 4.86GRR777 pKa = 11.84MADD780 pKa = 3.72DD781 pKa = 4.28FDD783 pKa = 3.83GLRR786 pKa = 11.84YY787 pKa = 9.29IASNADD793 pKa = 3.99LIVALGDD800 pKa = 4.28DD801 pKa = 4.1DD802 pKa = 5.53DD803 pKa = 4.39AAARR807 pKa = 11.84HH808 pKa = 6.02YY809 pKa = 10.69ILAGQQEE816 pKa = 4.5GRR818 pKa = 11.84DD819 pKa = 3.52VDD821 pKa = 4.41GFDD824 pKa = 3.54ALAYY828 pKa = 10.06AAANPDD834 pKa = 3.09LAAAFGSDD842 pKa = 2.95VDD844 pKa = 4.76ALTEE848 pKa = 4.16HH849 pKa = 6.83YY850 pKa = 10.33IDD852 pKa = 3.05TGYY855 pKa = 10.87FEE857 pKa = 5.13HH858 pKa = 7.26RR859 pKa = 11.84VVAPGADD866 pKa = 2.83MFMAGG871 pKa = 4.09
MM1 pKa = 8.13DD2 pKa = 3.97FTGGAGDD9 pKa = 5.82DD10 pKa = 4.37IYY12 pKa = 11.26TGDD15 pKa = 4.2NSDD18 pKa = 4.14EE19 pKa = 4.42YY20 pKa = 11.01ISGLGGNDD28 pKa = 3.01WLSGGEE34 pKa = 4.11GADD37 pKa = 3.92NIAGGAGADD46 pKa = 3.87RR47 pKa = 11.84LNGGDD52 pKa = 3.54GSDD55 pKa = 3.36VLFSFGRR62 pKa = 11.84TTNMMAPYY70 pKa = 9.6FPLLAGVVSDD80 pKa = 6.4DD81 pKa = 3.47IFADD85 pKa = 3.22VDD87 pKa = 3.97TLIGGAGDD95 pKa = 4.03DD96 pKa = 4.45FFFAGYY102 pKa = 10.04GDD104 pKa = 4.95HH105 pKa = 7.46IDD107 pKa = 3.78GGSYY111 pKa = 11.0DD112 pKa = 3.42SFGNRR117 pKa = 11.84LFISFQGATSGVVADD132 pKa = 6.13FRR134 pKa = 11.84PLQNGEE140 pKa = 4.37SITIGGGIISNIQNIGFLEE159 pKa = 4.11GRR161 pKa = 11.84EE162 pKa = 3.9YY163 pKa = 11.45DD164 pKa = 4.73DD165 pKa = 5.92FIASIDD171 pKa = 3.47TYY173 pKa = 10.32YY174 pKa = 10.57PSGADD179 pKa = 2.77IFGRR183 pKa = 11.84GGNDD187 pKa = 3.4TIIADD192 pKa = 4.39YY193 pKa = 10.58YY194 pKa = 10.4SGWGGAIWGGDD205 pKa = 3.22GDD207 pKa = 4.4DD208 pKa = 4.06TVDD211 pKa = 3.47ATGAQYY217 pKa = 10.78GARR220 pKa = 11.84VYY222 pKa = 11.18GEE224 pKa = 4.46AGNDD228 pKa = 3.82IIRR231 pKa = 11.84MNYY234 pKa = 7.29GTADD238 pKa = 3.59GGDD241 pKa = 3.89GDD243 pKa = 5.19DD244 pKa = 4.32IIYY247 pKa = 10.56GGSAFGGAGNDD258 pKa = 3.33RR259 pKa = 11.84LIDD262 pKa = 3.73VMSGDD267 pKa = 4.21GGDD270 pKa = 3.82GNDD273 pKa = 3.83IITLNFSYY281 pKa = 10.55YY282 pKa = 10.72GNGVATGGRR291 pKa = 11.84GDD293 pKa = 3.86DD294 pKa = 4.71LIRR297 pKa = 11.84GTDD300 pKa = 3.52YY301 pKa = 11.56GSVLVGGEE309 pKa = 4.09GADD312 pKa = 3.63EE313 pKa = 4.41LFGGAGADD321 pKa = 4.17LIYY324 pKa = 10.7TGGRR328 pKa = 11.84NEE330 pKa = 5.18DD331 pKa = 3.35GSALADD337 pKa = 3.36VGVEE341 pKa = 3.85KK342 pKa = 11.08DD343 pKa = 3.98FVSAGAGADD352 pKa = 3.96TIWAGVGDD360 pKa = 4.83DD361 pKa = 3.83VDD363 pKa = 5.12GGDD366 pKa = 4.5GADD369 pKa = 3.06TLYY372 pKa = 11.19YY373 pKa = 10.59SFGGATTGVDD383 pKa = 2.89ISTHH387 pKa = 4.67QFLGATARR395 pKa = 11.84VIGGGTIISIEE406 pKa = 4.09SLAEE410 pKa = 3.75LRR412 pKa = 11.84ASSFDD417 pKa = 3.53DD418 pKa = 4.27RR419 pKa = 11.84IVATTQDD426 pKa = 3.5VRR428 pKa = 11.84LSIYY432 pKa = 10.48GGAGNDD438 pKa = 3.42VVTSSGSSITFRR450 pKa = 11.84GEE452 pKa = 3.59EE453 pKa = 4.09GDD455 pKa = 4.07DD456 pKa = 3.4RR457 pKa = 11.84LISGEE462 pKa = 4.0AADD465 pKa = 4.02MFVGGFGTDD474 pKa = 3.25TIDD477 pKa = 3.4YY478 pKa = 10.37SLYY481 pKa = 8.9TNGVTVTLGLSGAIGIGGGGDD502 pKa = 3.43SLISVEE508 pKa = 4.3NVVGSAFSDD517 pKa = 4.11DD518 pKa = 3.45IQGNEE523 pKa = 4.29LANMLEE529 pKa = 4.46GGAGNDD535 pKa = 4.28TIHH538 pKa = 6.96GGAGDD543 pKa = 4.43DD544 pKa = 3.67RR545 pKa = 11.84LLPGTGHH552 pKa = 6.81NIVDD556 pKa = 4.08GGDD559 pKa = 3.65GFDD562 pKa = 3.6TLVLTGGVASYY573 pKa = 11.23NYY575 pKa = 9.56LVSGASTFVIGEE587 pKa = 4.17EE588 pKa = 4.2GASRR592 pKa = 11.84IVNVEE597 pKa = 3.7HH598 pKa = 6.59VAFANGALGSADD610 pKa = 3.46LAGSLSAFDD619 pKa = 3.73GLRR622 pKa = 11.84YY623 pKa = 8.98IAGYY627 pKa = 10.23GDD629 pKa = 5.42LIAAFGSDD637 pKa = 3.51ADD639 pKa = 3.95SATAHH644 pKa = 5.63YY645 pKa = 9.48VASGFAEE652 pKa = 4.24GRR654 pKa = 11.84DD655 pKa = 3.32AKK657 pKa = 10.91EE658 pKa = 3.92FDD660 pKa = 3.67VLDD663 pKa = 4.37YY664 pKa = 11.01IAGYY668 pKa = 10.5DD669 pKa = 3.71DD670 pKa = 6.12LIAAFGTDD678 pKa = 3.18TQAATSHH685 pKa = 6.3YY686 pKa = 9.32IDD688 pKa = 3.91GGFVEE693 pKa = 5.02GRR695 pKa = 11.84SDD697 pKa = 3.7ALFDD701 pKa = 3.64GLQYY705 pKa = 10.23IASYY709 pKa = 10.92GDD711 pKa = 3.93LIEE714 pKa = 5.77AFGTDD719 pKa = 2.83ADD721 pKa = 3.49AGARR725 pKa = 11.84HH726 pKa = 6.8FIQGGRR732 pKa = 11.84DD733 pKa = 3.2EE734 pKa = 5.16GRR736 pKa = 11.84SDD738 pKa = 4.61DD739 pKa = 5.51LFDD742 pKa = 3.91GLQYY746 pKa = 10.2VASYY750 pKa = 10.81GDD752 pKa = 4.23LIGAIGTDD760 pKa = 3.27VEE762 pKa = 4.28AAAEE766 pKa = 4.05HH767 pKa = 6.67FIKK770 pKa = 10.71NGYY773 pKa = 9.35GEE775 pKa = 4.86GRR777 pKa = 11.84MADD780 pKa = 3.72DD781 pKa = 4.28FDD783 pKa = 3.83GLRR786 pKa = 11.84YY787 pKa = 9.29IASNADD793 pKa = 3.99LIVALGDD800 pKa = 4.28DD801 pKa = 4.1DD802 pKa = 5.53DD803 pKa = 4.39AAARR807 pKa = 11.84HH808 pKa = 6.02YY809 pKa = 10.69ILAGQQEE816 pKa = 4.5GRR818 pKa = 11.84DD819 pKa = 3.52VDD821 pKa = 4.41GFDD824 pKa = 3.54ALAYY828 pKa = 10.06AAANPDD834 pKa = 3.09LAAAFGSDD842 pKa = 2.95VDD844 pKa = 4.76ALTEE848 pKa = 4.16HH849 pKa = 6.83YY850 pKa = 10.33IDD852 pKa = 3.05TGYY855 pKa = 10.87FEE857 pKa = 5.13HH858 pKa = 7.26RR859 pKa = 11.84VVAPGADD866 pKa = 2.83MFMAGG871 pKa = 4.09
Molecular weight: 88.93 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B3MRB7|A0A1B3MRB7_9SPHN Uncharacterized protein OS=Sphingobium sp. RAC03 OX=1843368 GN=BSY17_494 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.33RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 5.19GYY18 pKa = 7.59RR19 pKa = 11.84ARR21 pKa = 11.84MATPGGRR28 pKa = 11.84NIIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.33RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 5.19GYY18 pKa = 7.59RR19 pKa = 11.84ARR21 pKa = 11.84MATPGGRR28 pKa = 11.84NIIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1290787 |
29 |
2504 |
314.1 |
33.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.085 ± 0.057 | 0.819 ± 0.012 |
6.323 ± 0.033 | 5.002 ± 0.038 |
3.485 ± 0.023 | 8.8 ± 0.054 |
2.044 ± 0.02 | 5.295 ± 0.023 |
3.06 ± 0.029 | 9.797 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.643 ± 0.018 | 2.564 ± 0.025 |
5.307 ± 0.03 | 3.357 ± 0.024 |
7.191 ± 0.034 | 5.224 ± 0.027 |
5.33 ± 0.026 | 6.956 ± 0.024 |
1.442 ± 0.014 | 2.277 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
