
Plasmodiophora brassicae (Clubroot disease)
Taxonomy: cellular organisms; Eukaryota; Sar; Rhizaria; Endomyxa; Phytomyxea; Plasmodiophorida; Plasmodiophoridae; Plasmodiophora
Average proteome isoelectric point is 6.79
Get precalculated fractions of proteins
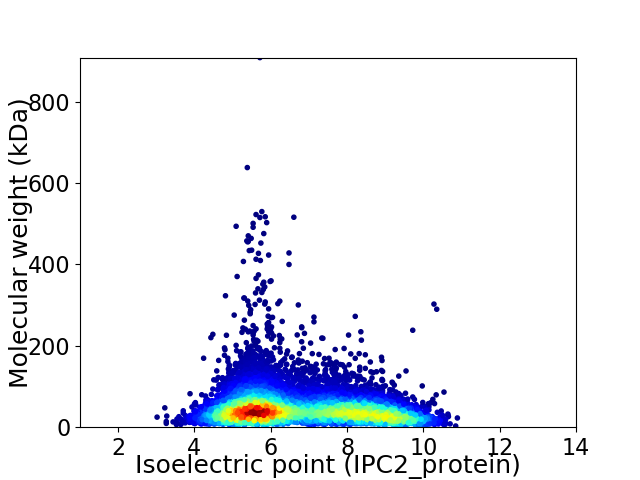
Virtual 2D-PAGE plot for 9720 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0G4ISR1|A0A0G4ISR1_PLABS Uncharacterized protein OS=Plasmodiophora brassicae OX=37360 GN=PBRA_006245 PE=4 SV=1
MM1 pKa = 7.61YY2 pKa = 7.89DD3 pKa = 3.26TPEE6 pKa = 4.38FGDD9 pKa = 3.58DD10 pKa = 3.91RR11 pKa = 11.84EE12 pKa = 4.65DD13 pKa = 3.46VSSGDD18 pKa = 3.58DD19 pKa = 3.59SVDD22 pKa = 3.24EE23 pKa = 4.31RR24 pKa = 11.84AVDD27 pKa = 4.17CGSDD31 pKa = 3.77DD32 pKa = 5.55DD33 pKa = 5.97DD34 pKa = 5.05SEE36 pKa = 4.94NYY38 pKa = 10.87GDD40 pKa = 4.33FEE42 pKa = 4.89VVGPADD48 pKa = 3.55QLTPTNPALAVVIEE62 pKa = 4.16AAEE65 pKa = 3.95QGDD68 pKa = 3.84VDD70 pKa = 3.97RR71 pKa = 11.84LEE73 pKa = 4.76NGLSALTTSVNCVGDD88 pKa = 4.42DD89 pKa = 3.78GDD91 pKa = 4.34SALHH95 pKa = 5.87LASLFGHH102 pKa = 6.06LQCVEE107 pKa = 4.31LLLSHH112 pKa = 6.62GAEE115 pKa = 5.08ASFTDD120 pKa = 4.31CNGGLALHH128 pKa = 6.91DD129 pKa = 4.2ACASGHH135 pKa = 6.13LDD137 pKa = 3.1IVKK140 pKa = 10.15VLLKK144 pKa = 10.39SAPHH148 pKa = 6.21TIGVIDD154 pKa = 4.54EE155 pKa = 5.54DD156 pKa = 4.73GDD158 pKa = 4.49SPLHH162 pKa = 5.26NAARR166 pKa = 11.84GNHH169 pKa = 6.0LDD171 pKa = 3.27IVSFLLDD178 pKa = 3.34NGARR182 pKa = 11.84SCLEE186 pKa = 3.52IANLEE191 pKa = 4.22GEE193 pKa = 4.6LPAMCTTCPKK203 pKa = 10.0VRR205 pKa = 11.84SLLTHH210 pKa = 5.38QTPAPP215 pKa = 3.73
MM1 pKa = 7.61YY2 pKa = 7.89DD3 pKa = 3.26TPEE6 pKa = 4.38FGDD9 pKa = 3.58DD10 pKa = 3.91RR11 pKa = 11.84EE12 pKa = 4.65DD13 pKa = 3.46VSSGDD18 pKa = 3.58DD19 pKa = 3.59SVDD22 pKa = 3.24EE23 pKa = 4.31RR24 pKa = 11.84AVDD27 pKa = 4.17CGSDD31 pKa = 3.77DD32 pKa = 5.55DD33 pKa = 5.97DD34 pKa = 5.05SEE36 pKa = 4.94NYY38 pKa = 10.87GDD40 pKa = 4.33FEE42 pKa = 4.89VVGPADD48 pKa = 3.55QLTPTNPALAVVIEE62 pKa = 4.16AAEE65 pKa = 3.95QGDD68 pKa = 3.84VDD70 pKa = 3.97RR71 pKa = 11.84LEE73 pKa = 4.76NGLSALTTSVNCVGDD88 pKa = 4.42DD89 pKa = 3.78GDD91 pKa = 4.34SALHH95 pKa = 5.87LASLFGHH102 pKa = 6.06LQCVEE107 pKa = 4.31LLLSHH112 pKa = 6.62GAEE115 pKa = 5.08ASFTDD120 pKa = 4.31CNGGLALHH128 pKa = 6.91DD129 pKa = 4.2ACASGHH135 pKa = 6.13LDD137 pKa = 3.1IVKK140 pKa = 10.15VLLKK144 pKa = 10.39SAPHH148 pKa = 6.21TIGVIDD154 pKa = 4.54EE155 pKa = 5.54DD156 pKa = 4.73GDD158 pKa = 4.49SPLHH162 pKa = 5.26NAARR166 pKa = 11.84GNHH169 pKa = 6.0LDD171 pKa = 3.27IVSFLLDD178 pKa = 3.34NGARR182 pKa = 11.84SCLEE186 pKa = 3.52IANLEE191 pKa = 4.22GEE193 pKa = 4.6LPAMCTTCPKK203 pKa = 10.0VRR205 pKa = 11.84SLLTHH210 pKa = 5.38QTPAPP215 pKa = 3.73
Molecular weight: 22.53 kDa
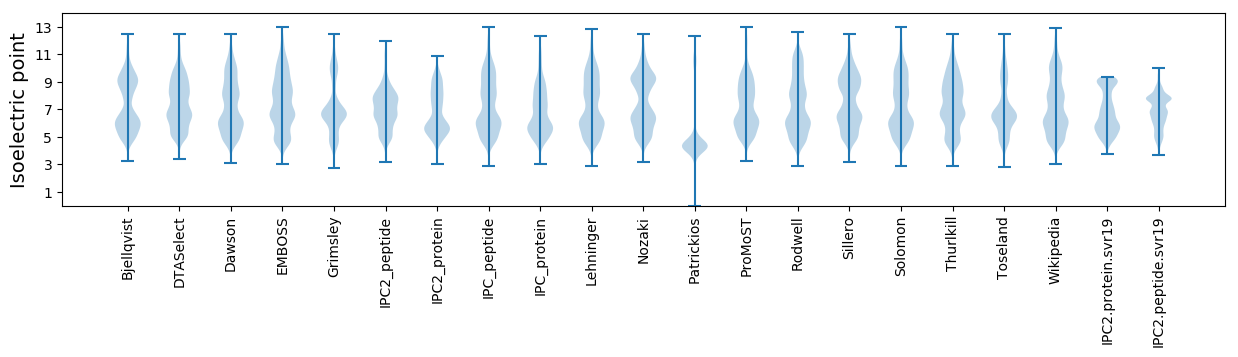
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0G4IMF8|A0A0G4IMF8_PLABS Uncharacterized protein OS=Plasmodiophora brassicae OX=37360 GN=PBRA_005102 PE=4 SV=1
MM1 pKa = 7.41RR2 pKa = 11.84LAWVVVLLATAVLTGPLQALIDD24 pKa = 4.23GVTPQQVVDD33 pKa = 4.56LLNSIRR39 pKa = 11.84GNVVPTAAQMVRR51 pKa = 11.84LTWNDD56 pKa = 3.1TLAAAAARR64 pKa = 11.84YY65 pKa = 8.01AATQCAANFDD75 pKa = 4.29GQGPQCPDD83 pKa = 3.03CANWIGEE90 pKa = 4.01GMYY93 pKa = 10.47PSATTHH99 pKa = 6.14GMLDD103 pKa = 3.83SINDD107 pKa = 3.59LMGEE111 pKa = 4.14KK112 pKa = 10.18AQWSCTQSGTTLSCKK127 pKa = 9.99CSSYY131 pKa = 10.8HH132 pKa = 7.44CYY134 pKa = 10.94DD135 pKa = 2.88NWKK138 pKa = 10.68VILNDD143 pKa = 3.29GATTVGCGEE152 pKa = 4.83LLASTCGGSYY162 pKa = 10.6NIYY165 pKa = 9.19VCAFNGFSSYY175 pKa = 10.65AHH177 pKa = 7.71PYY179 pKa = 8.47TLSATNSSCTQGTCPSAYY197 pKa = 8.76PSCVNSLCTRR207 pKa = 11.84RR208 pKa = 11.84PATQFTTRR216 pKa = 11.84RR217 pKa = 11.84TSVTKK222 pKa = 10.03MRR224 pKa = 11.84TKK226 pKa = 9.76RR227 pKa = 11.84TSVKK231 pKa = 10.18QLTKK235 pKa = 10.38RR236 pKa = 11.84RR237 pKa = 11.84TSVKK241 pKa = 10.01QLTKK245 pKa = 10.38RR246 pKa = 11.84RR247 pKa = 11.84TSVKK251 pKa = 10.01QLTKK255 pKa = 10.45RR256 pKa = 11.84RR257 pKa = 11.84TSVTTLRR264 pKa = 11.84ATLQRR269 pKa = 11.84ANVPSLRR276 pKa = 11.84KK277 pKa = 8.6RR278 pKa = 11.84TSRR281 pKa = 11.84PATGHH286 pKa = 4.27VKK288 pKa = 10.29RR289 pKa = 11.84RR290 pKa = 11.84TSTMRR295 pKa = 11.84PRR297 pKa = 11.84SRR299 pKa = 11.84LTTTPRR305 pKa = 11.84LLANSSGVSTRR316 pKa = 11.84SLEE319 pKa = 4.02PVRR322 pKa = 11.84QGSVRR327 pKa = 11.84SFVTSLGTGMAVTTPRR343 pKa = 11.84SKK345 pKa = 10.58RR346 pKa = 11.84SSSKK350 pKa = 11.01GPGADD355 pKa = 3.22TRR357 pKa = 11.84SASAPRR363 pKa = 11.84SPSLFSDD370 pKa = 3.76VSDD373 pKa = 3.27TTLFVVGSVAVVCMAALALLIRR395 pKa = 11.84RR396 pKa = 11.84RR397 pKa = 11.84NQQEE401 pKa = 3.54LSARR405 pKa = 11.84AIPEE409 pKa = 4.49FINWPTDD416 pKa = 3.48PLATSSVQSVGQSAFTGDD434 pKa = 3.94TRR436 pKa = 11.84PADD439 pKa = 4.19PPAHH443 pKa = 7.0DD444 pKa = 5.33PFNAMPKK451 pKa = 9.88AVRR454 pKa = 11.84RR455 pKa = 11.84LQARR459 pKa = 11.84GVARR463 pKa = 11.84DD464 pKa = 3.86KK465 pKa = 11.11FNQGARR471 pKa = 11.84TVSDD475 pKa = 3.82PSEE478 pKa = 4.51SDD480 pKa = 2.94AA481 pKa = 5.34
MM1 pKa = 7.41RR2 pKa = 11.84LAWVVVLLATAVLTGPLQALIDD24 pKa = 4.23GVTPQQVVDD33 pKa = 4.56LLNSIRR39 pKa = 11.84GNVVPTAAQMVRR51 pKa = 11.84LTWNDD56 pKa = 3.1TLAAAAARR64 pKa = 11.84YY65 pKa = 8.01AATQCAANFDD75 pKa = 4.29GQGPQCPDD83 pKa = 3.03CANWIGEE90 pKa = 4.01GMYY93 pKa = 10.47PSATTHH99 pKa = 6.14GMLDD103 pKa = 3.83SINDD107 pKa = 3.59LMGEE111 pKa = 4.14KK112 pKa = 10.18AQWSCTQSGTTLSCKK127 pKa = 9.99CSSYY131 pKa = 10.8HH132 pKa = 7.44CYY134 pKa = 10.94DD135 pKa = 2.88NWKK138 pKa = 10.68VILNDD143 pKa = 3.29GATTVGCGEE152 pKa = 4.83LLASTCGGSYY162 pKa = 10.6NIYY165 pKa = 9.19VCAFNGFSSYY175 pKa = 10.65AHH177 pKa = 7.71PYY179 pKa = 8.47TLSATNSSCTQGTCPSAYY197 pKa = 8.76PSCVNSLCTRR207 pKa = 11.84RR208 pKa = 11.84PATQFTTRR216 pKa = 11.84RR217 pKa = 11.84TSVTKK222 pKa = 10.03MRR224 pKa = 11.84TKK226 pKa = 9.76RR227 pKa = 11.84TSVKK231 pKa = 10.18QLTKK235 pKa = 10.38RR236 pKa = 11.84RR237 pKa = 11.84TSVKK241 pKa = 10.01QLTKK245 pKa = 10.38RR246 pKa = 11.84RR247 pKa = 11.84TSVKK251 pKa = 10.01QLTKK255 pKa = 10.45RR256 pKa = 11.84RR257 pKa = 11.84TSVTTLRR264 pKa = 11.84ATLQRR269 pKa = 11.84ANVPSLRR276 pKa = 11.84KK277 pKa = 8.6RR278 pKa = 11.84TSRR281 pKa = 11.84PATGHH286 pKa = 4.27VKK288 pKa = 10.29RR289 pKa = 11.84RR290 pKa = 11.84TSTMRR295 pKa = 11.84PRR297 pKa = 11.84SRR299 pKa = 11.84LTTTPRR305 pKa = 11.84LLANSSGVSTRR316 pKa = 11.84SLEE319 pKa = 4.02PVRR322 pKa = 11.84QGSVRR327 pKa = 11.84SFVTSLGTGMAVTTPRR343 pKa = 11.84SKK345 pKa = 10.58RR346 pKa = 11.84SSSKK350 pKa = 11.01GPGADD355 pKa = 3.22TRR357 pKa = 11.84SASAPRR363 pKa = 11.84SPSLFSDD370 pKa = 3.76VSDD373 pKa = 3.27TTLFVVGSVAVVCMAALALLIRR395 pKa = 11.84RR396 pKa = 11.84RR397 pKa = 11.84NQQEE401 pKa = 3.54LSARR405 pKa = 11.84AIPEE409 pKa = 4.49FINWPTDD416 pKa = 3.48PLATSSVQSVGQSAFTGDD434 pKa = 3.94TRR436 pKa = 11.84PADD439 pKa = 4.19PPAHH443 pKa = 7.0DD444 pKa = 5.33PFNAMPKK451 pKa = 9.88AVRR454 pKa = 11.84RR455 pKa = 11.84LQARR459 pKa = 11.84GVARR463 pKa = 11.84DD464 pKa = 3.86KK465 pKa = 11.11FNQGARR471 pKa = 11.84TVSDD475 pKa = 3.82PSEE478 pKa = 4.51SDD480 pKa = 2.94AA481 pKa = 5.34
Molecular weight: 51.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4369470 |
23 |
8637 |
449.5 |
49.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.297 ± 0.031 | 1.859 ± 0.015 |
6.347 ± 0.023 | 4.919 ± 0.024 |
3.66 ± 0.017 | 6.112 ± 0.021 |
2.517 ± 0.011 | 4.573 ± 0.014 |
3.564 ± 0.02 | 9.544 ± 0.029 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.375 ± 0.011 | 3.024 ± 0.011 |
5.319 ± 0.024 | 3.885 ± 0.015 |
7.378 ± 0.028 | 7.755 ± 0.029 |
5.458 ± 0.02 | 7.912 ± 0.019 |
1.245 ± 0.009 | 2.249 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
