
Burdock mottle virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Hepelivirales; Benyviridae; Benyvirus
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
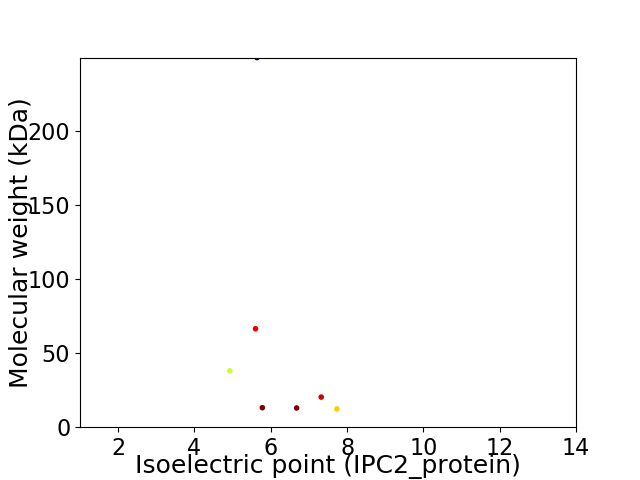
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|S6B9F8|S6B9F8_9VIRU Coat protein read-through product OS=Burdock mottle virus OX=1324959 GN=CP-RT PE=4 SV=1
MM1 pKa = 7.33TEE3 pKa = 3.65WASEE7 pKa = 4.02NPHH10 pKa = 6.92DD11 pKa = 4.19FFSVLEE17 pKa = 4.28RR18 pKa = 11.84NCGNAGFSWTGVRR31 pKa = 11.84PRR33 pKa = 11.84TITYY37 pKa = 10.19SDD39 pKa = 3.84LMSSGALVNLQSLLEE54 pKa = 4.22SEE56 pKa = 4.74SFSGCVRR63 pKa = 11.84SGDD66 pKa = 3.39IAAVKK71 pKa = 10.37SDD73 pKa = 3.89VVSKK77 pKa = 10.6MDD79 pKa = 3.56GKK81 pKa = 10.4DD82 pKa = 2.86WQARR86 pKa = 11.84CGLVTGVAGSGKK98 pKa = 8.53STLIKK103 pKa = 9.88TLLTSGEE110 pKa = 4.02GRR112 pKa = 11.84VVLGLPNSSLLKK124 pKa = 10.61GVFSGCPNAFLIDD137 pKa = 3.92DD138 pKa = 5.1LFTSEE143 pKa = 4.33IHH145 pKa = 5.21LQRR148 pKa = 11.84YY149 pKa = 5.38QTMLVDD155 pKa = 4.41EE156 pKa = 4.65FTKK159 pKa = 10.73VHH161 pKa = 5.5MCEE164 pKa = 4.24VMCLCVLLGVKK175 pKa = 10.29NLVCFGDD182 pKa = 4.8FSQSLNYY189 pKa = 9.86KK190 pKa = 9.56AGSVVNYY197 pKa = 8.82GLPVLAKK204 pKa = 9.92SDD206 pKa = 3.3TSKK209 pKa = 11.15RR210 pKa = 11.84FGKK213 pKa = 10.31KK214 pKa = 8.89IAGLMSGSGCGNVRR228 pKa = 11.84GSDD231 pKa = 3.6SVNDD235 pKa = 3.82DD236 pKa = 3.37VSFEE240 pKa = 4.22DD241 pKa = 5.07LMGKK245 pKa = 9.32LRR247 pKa = 11.84DD248 pKa = 3.61MSTVLVASEE257 pKa = 4.28EE258 pKa = 4.46SQKK261 pKa = 10.94EE262 pKa = 3.91LADD265 pKa = 3.87CDD267 pKa = 3.32IDD269 pKa = 4.03SFLWSEE275 pKa = 4.35VQGQTFDD282 pKa = 3.38VVEE285 pKa = 4.11VVLYY289 pKa = 10.72DD290 pKa = 3.73EE291 pKa = 5.58YY292 pKa = 11.42DD293 pKa = 3.62DD294 pKa = 6.51KK295 pKa = 11.62LICDD299 pKa = 3.74SNIRR303 pKa = 11.84TVLLSRR309 pKa = 11.84ARR311 pKa = 11.84KK312 pKa = 9.75CNVLRR317 pKa = 11.84FGPNIRR323 pKa = 11.84ARR325 pKa = 11.84FEE327 pKa = 4.22SGNFGCGGNDD337 pKa = 2.91SSYY340 pKa = 11.57SGDD343 pKa = 3.38TLRR346 pKa = 11.84EE347 pKa = 3.86EE348 pKa = 4.42RR349 pKa = 4.46
MM1 pKa = 7.33TEE3 pKa = 3.65WASEE7 pKa = 4.02NPHH10 pKa = 6.92DD11 pKa = 4.19FFSVLEE17 pKa = 4.28RR18 pKa = 11.84NCGNAGFSWTGVRR31 pKa = 11.84PRR33 pKa = 11.84TITYY37 pKa = 10.19SDD39 pKa = 3.84LMSSGALVNLQSLLEE54 pKa = 4.22SEE56 pKa = 4.74SFSGCVRR63 pKa = 11.84SGDD66 pKa = 3.39IAAVKK71 pKa = 10.37SDD73 pKa = 3.89VVSKK77 pKa = 10.6MDD79 pKa = 3.56GKK81 pKa = 10.4DD82 pKa = 2.86WQARR86 pKa = 11.84CGLVTGVAGSGKK98 pKa = 8.53STLIKK103 pKa = 9.88TLLTSGEE110 pKa = 4.02GRR112 pKa = 11.84VVLGLPNSSLLKK124 pKa = 10.61GVFSGCPNAFLIDD137 pKa = 3.92DD138 pKa = 5.1LFTSEE143 pKa = 4.33IHH145 pKa = 5.21LQRR148 pKa = 11.84YY149 pKa = 5.38QTMLVDD155 pKa = 4.41EE156 pKa = 4.65FTKK159 pKa = 10.73VHH161 pKa = 5.5MCEE164 pKa = 4.24VMCLCVLLGVKK175 pKa = 10.29NLVCFGDD182 pKa = 4.8FSQSLNYY189 pKa = 9.86KK190 pKa = 9.56AGSVVNYY197 pKa = 8.82GLPVLAKK204 pKa = 9.92SDD206 pKa = 3.3TSKK209 pKa = 11.15RR210 pKa = 11.84FGKK213 pKa = 10.31KK214 pKa = 8.89IAGLMSGSGCGNVRR228 pKa = 11.84GSDD231 pKa = 3.6SVNDD235 pKa = 3.82DD236 pKa = 3.37VSFEE240 pKa = 4.22DD241 pKa = 5.07LMGKK245 pKa = 9.32LRR247 pKa = 11.84DD248 pKa = 3.61MSTVLVASEE257 pKa = 4.28EE258 pKa = 4.46SQKK261 pKa = 10.94EE262 pKa = 3.91LADD265 pKa = 3.87CDD267 pKa = 3.32IDD269 pKa = 4.03SFLWSEE275 pKa = 4.35VQGQTFDD282 pKa = 3.38VVEE285 pKa = 4.11VVLYY289 pKa = 10.72DD290 pKa = 3.73EE291 pKa = 5.58YY292 pKa = 11.42DD293 pKa = 3.62DD294 pKa = 6.51KK295 pKa = 11.62LICDD299 pKa = 3.74SNIRR303 pKa = 11.84TVLLSRR309 pKa = 11.84ARR311 pKa = 11.84KK312 pKa = 9.75CNVLRR317 pKa = 11.84FGPNIRR323 pKa = 11.84ARR325 pKa = 11.84FEE327 pKa = 4.22SGNFGCGGNDD337 pKa = 2.91SSYY340 pKa = 11.57SGDD343 pKa = 3.38TLRR346 pKa = 11.84EE347 pKa = 3.86EE348 pKa = 4.42RR349 pKa = 4.46
Molecular weight: 37.99 kDa
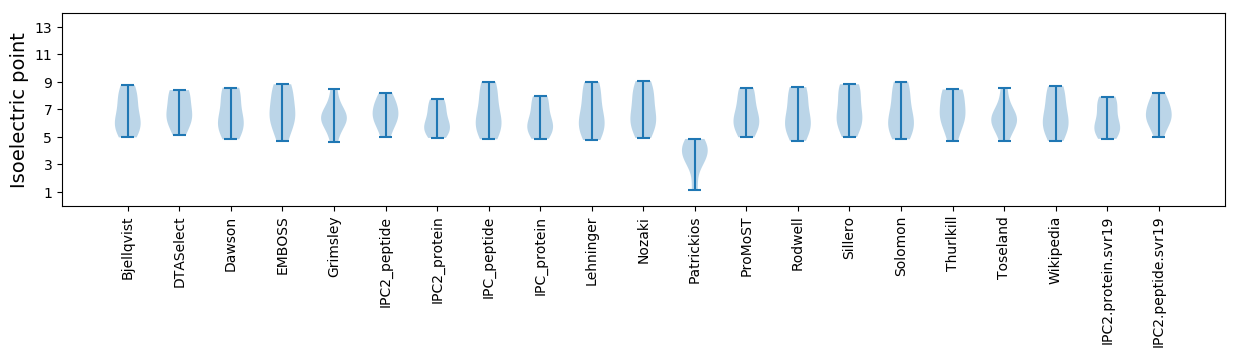
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S6C3R5|S6C3R5_9VIRU Capsid protein OS=Burdock mottle virus OX=1324959 GN=CP PE=4 SV=1
MM1 pKa = 6.86TRR3 pKa = 11.84EE4 pKa = 3.39IRR6 pKa = 11.84ARR8 pKa = 11.84EE9 pKa = 4.17SNTKK13 pKa = 10.34YY14 pKa = 10.17IVLGVCVVAFICFLGFSQQKK34 pKa = 9.64HH35 pKa = 4.69ATHH38 pKa = 6.54SGDD41 pKa = 3.28GVGVPRR47 pKa = 11.84FANGGSYY54 pKa = 10.5RR55 pKa = 11.84DD56 pKa = 3.89GTRR59 pKa = 11.84SMNFNSNNPNAYY71 pKa = 8.42GCKK74 pKa = 9.7SEE76 pKa = 4.62GFFGGFEE83 pKa = 3.98KK84 pKa = 10.85LALLFLVLGIILYY97 pKa = 9.96VAGGCAGGDD106 pKa = 3.58HH107 pKa = 6.4VCNGGCCKK115 pKa = 10.3VV116 pKa = 3.14
MM1 pKa = 6.86TRR3 pKa = 11.84EE4 pKa = 3.39IRR6 pKa = 11.84ARR8 pKa = 11.84EE9 pKa = 4.17SNTKK13 pKa = 10.34YY14 pKa = 10.17IVLGVCVVAFICFLGFSQQKK34 pKa = 9.64HH35 pKa = 4.69ATHH38 pKa = 6.54SGDD41 pKa = 3.28GVGVPRR47 pKa = 11.84FANGGSYY54 pKa = 10.5RR55 pKa = 11.84DD56 pKa = 3.89GTRR59 pKa = 11.84SMNFNSNNPNAYY71 pKa = 8.42GCKK74 pKa = 9.7SEE76 pKa = 4.62GFFGGFEE83 pKa = 3.98KK84 pKa = 10.85LALLFLVLGIILYY97 pKa = 9.96VAGGCAGGDD106 pKa = 3.58HH107 pKa = 6.4VCNGGCCKK115 pKa = 10.3VV116 pKa = 3.14
Molecular weight: 12.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3711 |
116 |
2217 |
530.1 |
58.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.114 ± 0.945 | 1.967 ± 0.614 |
6.117 ± 0.379 | 5.901 ± 0.482 |
5.012 ± 0.228 | 6.871 ± 0.949 |
2.129 ± 0.213 | 4.554 ± 0.436 |
5.74 ± 0.415 | 9.054 ± 0.396 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.506 ± 0.263 | 3.961 ± 0.248 |
3.719 ± 0.407 | 3.288 ± 0.42 |
5.039 ± 0.186 | 8.946 ± 0.581 |
5.147 ± 0.238 | 8.569 ± 1.053 |
1.293 ± 0.211 | 3.045 ± 0.405 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
