
Tobacco mottle leaf curl virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Begomovirus
Average proteome isoelectric point is 8.07
Get precalculated fractions of proteins
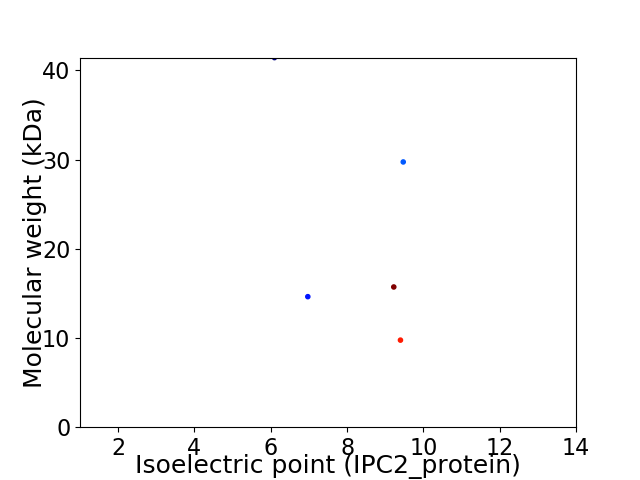
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B2ZWE7|B2ZWE7_9GEMI AC4 protein OS=Tobacco mottle leaf curl virus OX=530536 GN=AC4 PE=3 SV=1
MM1 pKa = 7.64PSVKK5 pKa = 10.02KK6 pKa = 10.46FRR8 pKa = 11.84VQSRR12 pKa = 11.84NYY14 pKa = 9.52FLTYY18 pKa = 8.6PQCSLSKK25 pKa = 11.0EE26 pKa = 4.09EE27 pKa = 5.81ALSQLQNLDD36 pKa = 3.35TPVNKK41 pKa = 10.11KK42 pKa = 9.98FIKK45 pKa = 9.67ICRR48 pKa = 11.84EE49 pKa = 3.52LHH51 pKa = 6.26EE52 pKa = 4.93NGEE55 pKa = 4.29PHH57 pKa = 6.86LHH59 pKa = 5.98VLVQFEE65 pKa = 4.35GKK67 pKa = 9.34YY68 pKa = 9.62VCTNYY73 pKa = 10.47RR74 pKa = 11.84FFDD77 pKa = 3.75LVSPSRR83 pKa = 11.84SAHH86 pKa = 5.01FHH88 pKa = 6.64PNVQGAKK95 pKa = 9.76SSSDD99 pKa = 3.11VKK101 pKa = 11.24SYY103 pKa = 10.79IDD105 pKa = 3.7KK106 pKa = 11.33DD107 pKa = 3.49GDD109 pKa = 3.67TLEE112 pKa = 4.1WGEE115 pKa = 3.95FQVDD119 pKa = 3.21GRR121 pKa = 11.84SARR124 pKa = 11.84GGCQSANDD132 pKa = 4.17TYY134 pKa = 11.77AKK136 pKa = 10.57ALNASSAEE144 pKa = 4.0EE145 pKa = 3.51ALQIIKK151 pKa = 10.28EE152 pKa = 4.13EE153 pKa = 4.13QPQHH157 pKa = 6.73FFLQHH162 pKa = 6.07HH163 pKa = 6.14NLVANAQRR171 pKa = 11.84IFQKK175 pKa = 10.74APEE178 pKa = 4.11PWVPPFQLSSLTNVPEE194 pKa = 4.61EE195 pKa = 4.01MQEE198 pKa = 3.79WADD201 pKa = 3.73EE202 pKa = 4.25YY203 pKa = 11.01FGRR206 pKa = 11.84GSAARR211 pKa = 11.84PEE213 pKa = 4.21RR214 pKa = 11.84PMSIIVEE221 pKa = 4.27GDD223 pKa = 3.04SRR225 pKa = 11.84TGKK228 pKa = 8.49TMWACSKK235 pKa = 9.46TMWARR240 pKa = 11.84ALGPHH245 pKa = 6.63NYY247 pKa = 10.14LSGHH251 pKa = 6.96LDD253 pKa = 3.47FNHH256 pKa = 6.23RR257 pKa = 11.84VYY259 pKa = 11.3SNEE262 pKa = 3.44VEE264 pKa = 4.32YY265 pKa = 11.37NVIDD269 pKa = 4.4DD270 pKa = 4.14VAPHH274 pKa = 5.29YY275 pKa = 10.97LKK277 pKa = 10.78LKK279 pKa = 9.14HH280 pKa = 5.88WKK282 pKa = 9.55EE283 pKa = 3.9LLGSQKK289 pKa = 10.72DD290 pKa = 3.48WQSNCKK296 pKa = 9.38YY297 pKa = 10.21GKK299 pKa = 9.18PVQIKK304 pKa = 9.93GGIPAIVLCNPGEE317 pKa = 4.16GASYY321 pKa = 11.29KK322 pKa = 10.87DD323 pKa = 3.72FLDD326 pKa = 3.9KK327 pKa = 11.36EE328 pKa = 4.72EE329 pKa = 4.4NSSLRR334 pKa = 11.84NFGDD338 pKa = 3.51RR339 pKa = 11.84CGSSPPAPLYY349 pKa = 10.65QEE351 pKa = 4.48GTQASQRR358 pKa = 11.84GGPSGGDD365 pKa = 3.25ALII368 pKa = 4.53
MM1 pKa = 7.64PSVKK5 pKa = 10.02KK6 pKa = 10.46FRR8 pKa = 11.84VQSRR12 pKa = 11.84NYY14 pKa = 9.52FLTYY18 pKa = 8.6PQCSLSKK25 pKa = 11.0EE26 pKa = 4.09EE27 pKa = 5.81ALSQLQNLDD36 pKa = 3.35TPVNKK41 pKa = 10.11KK42 pKa = 9.98FIKK45 pKa = 9.67ICRR48 pKa = 11.84EE49 pKa = 3.52LHH51 pKa = 6.26EE52 pKa = 4.93NGEE55 pKa = 4.29PHH57 pKa = 6.86LHH59 pKa = 5.98VLVQFEE65 pKa = 4.35GKK67 pKa = 9.34YY68 pKa = 9.62VCTNYY73 pKa = 10.47RR74 pKa = 11.84FFDD77 pKa = 3.75LVSPSRR83 pKa = 11.84SAHH86 pKa = 5.01FHH88 pKa = 6.64PNVQGAKK95 pKa = 9.76SSSDD99 pKa = 3.11VKK101 pKa = 11.24SYY103 pKa = 10.79IDD105 pKa = 3.7KK106 pKa = 11.33DD107 pKa = 3.49GDD109 pKa = 3.67TLEE112 pKa = 4.1WGEE115 pKa = 3.95FQVDD119 pKa = 3.21GRR121 pKa = 11.84SARR124 pKa = 11.84GGCQSANDD132 pKa = 4.17TYY134 pKa = 11.77AKK136 pKa = 10.57ALNASSAEE144 pKa = 4.0EE145 pKa = 3.51ALQIIKK151 pKa = 10.28EE152 pKa = 4.13EE153 pKa = 4.13QPQHH157 pKa = 6.73FFLQHH162 pKa = 6.07HH163 pKa = 6.14NLVANAQRR171 pKa = 11.84IFQKK175 pKa = 10.74APEE178 pKa = 4.11PWVPPFQLSSLTNVPEE194 pKa = 4.61EE195 pKa = 4.01MQEE198 pKa = 3.79WADD201 pKa = 3.73EE202 pKa = 4.25YY203 pKa = 11.01FGRR206 pKa = 11.84GSAARR211 pKa = 11.84PEE213 pKa = 4.21RR214 pKa = 11.84PMSIIVEE221 pKa = 4.27GDD223 pKa = 3.04SRR225 pKa = 11.84TGKK228 pKa = 8.49TMWACSKK235 pKa = 9.46TMWARR240 pKa = 11.84ALGPHH245 pKa = 6.63NYY247 pKa = 10.14LSGHH251 pKa = 6.96LDD253 pKa = 3.47FNHH256 pKa = 6.23RR257 pKa = 11.84VYY259 pKa = 11.3SNEE262 pKa = 3.44VEE264 pKa = 4.32YY265 pKa = 11.37NVIDD269 pKa = 4.4DD270 pKa = 4.14VAPHH274 pKa = 5.29YY275 pKa = 10.97LKK277 pKa = 10.78LKK279 pKa = 9.14HH280 pKa = 5.88WKK282 pKa = 9.55EE283 pKa = 3.9LLGSQKK289 pKa = 10.72DD290 pKa = 3.48WQSNCKK296 pKa = 9.38YY297 pKa = 10.21GKK299 pKa = 9.18PVQIKK304 pKa = 9.93GGIPAIVLCNPGEE317 pKa = 4.16GASYY321 pKa = 11.29KK322 pKa = 10.87DD323 pKa = 3.72FLDD326 pKa = 3.9KK327 pKa = 11.36EE328 pKa = 4.72EE329 pKa = 4.4NSSLRR334 pKa = 11.84NFGDD338 pKa = 3.51RR339 pKa = 11.84CGSSPPAPLYY349 pKa = 10.65QEE351 pKa = 4.48GTQASQRR358 pKa = 11.84GGPSGGDD365 pKa = 3.25ALII368 pKa = 4.53
Molecular weight: 41.43 kDa
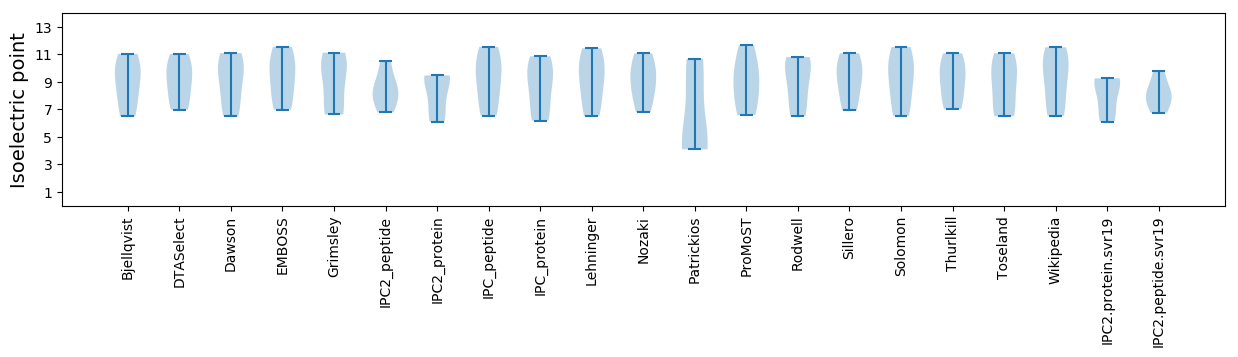
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B2ZWE4|B2ZWE4_9GEMI Replication enhancer OS=Tobacco mottle leaf curl virus OX=530536 GN=AC3 PE=3 SV=1
MM1 pKa = 8.04PKK3 pKa = 9.63RR4 pKa = 11.84DD5 pKa = 4.08LPWRR9 pKa = 11.84PYY11 pKa = 11.06AKK13 pKa = 9.39ISKK16 pKa = 9.56VSRR19 pKa = 11.84NANYY23 pKa = 10.37SPRR26 pKa = 11.84TRR28 pKa = 11.84VGPIVDD34 pKa = 4.98KK35 pKa = 10.94AAEE38 pKa = 4.1WVNRR42 pKa = 11.84PMYY45 pKa = 9.79RR46 pKa = 11.84KK47 pKa = 8.63PRR49 pKa = 11.84IYY51 pKa = 9.74RR52 pKa = 11.84TVRR55 pKa = 11.84TPDD58 pKa = 3.1VPRR61 pKa = 11.84GCEE64 pKa = 4.34GPCKK68 pKa = 9.32VQSYY72 pKa = 6.94EE73 pKa = 3.74QRR75 pKa = 11.84HH76 pKa = 6.38DD77 pKa = 3.16ISHH80 pKa = 6.13VGKK83 pKa = 10.55VMCISDD89 pKa = 3.56VTRR92 pKa = 11.84GNGITHH98 pKa = 6.78RR99 pKa = 11.84VGKK102 pKa = 9.44RR103 pKa = 11.84FCVKK107 pKa = 9.92SVYY110 pKa = 10.25ILGKK114 pKa = 9.79IGKK117 pKa = 8.34IWMDD121 pKa = 3.43EE122 pKa = 4.01NIKK125 pKa = 10.61LKK127 pKa = 10.73NHH129 pKa = 5.95TNSVMFWLVRR139 pKa = 11.84DD140 pKa = 3.7RR141 pKa = 11.84RR142 pKa = 11.84PYY144 pKa = 8.05GTPMDD149 pKa = 4.6FGQVFNMFDD158 pKa = 3.99NEE160 pKa = 4.11PSTATVKK167 pKa = 10.81NDD169 pKa = 2.84LRR171 pKa = 11.84DD172 pKa = 3.6RR173 pKa = 11.84YY174 pKa = 9.91QVMHH178 pKa = 7.14RR179 pKa = 11.84FHH181 pKa = 7.1AKK183 pKa = 8.01VTGGQYY189 pKa = 11.07ASNEE193 pKa = 3.78QAIVRR198 pKa = 11.84RR199 pKa = 11.84FWKK202 pKa = 10.31VNNHH206 pKa = 4.48VVYY209 pKa = 10.67NHH211 pKa = 5.8QEE213 pKa = 3.47AGKK216 pKa = 10.41YY217 pKa = 8.39EE218 pKa = 4.09NHH220 pKa = 6.52TEE222 pKa = 3.99NALLLYY228 pKa = 7.29MACTHH233 pKa = 7.07ASNPVYY239 pKa = 9.86ATLKK243 pKa = 9.47IRR245 pKa = 11.84IYY247 pKa = 10.66FYY249 pKa = 11.18DD250 pKa = 3.06SVMNN254 pKa = 4.52
MM1 pKa = 8.04PKK3 pKa = 9.63RR4 pKa = 11.84DD5 pKa = 4.08LPWRR9 pKa = 11.84PYY11 pKa = 11.06AKK13 pKa = 9.39ISKK16 pKa = 9.56VSRR19 pKa = 11.84NANYY23 pKa = 10.37SPRR26 pKa = 11.84TRR28 pKa = 11.84VGPIVDD34 pKa = 4.98KK35 pKa = 10.94AAEE38 pKa = 4.1WVNRR42 pKa = 11.84PMYY45 pKa = 9.79RR46 pKa = 11.84KK47 pKa = 8.63PRR49 pKa = 11.84IYY51 pKa = 9.74RR52 pKa = 11.84TVRR55 pKa = 11.84TPDD58 pKa = 3.1VPRR61 pKa = 11.84GCEE64 pKa = 4.34GPCKK68 pKa = 9.32VQSYY72 pKa = 6.94EE73 pKa = 3.74QRR75 pKa = 11.84HH76 pKa = 6.38DD77 pKa = 3.16ISHH80 pKa = 6.13VGKK83 pKa = 10.55VMCISDD89 pKa = 3.56VTRR92 pKa = 11.84GNGITHH98 pKa = 6.78RR99 pKa = 11.84VGKK102 pKa = 9.44RR103 pKa = 11.84FCVKK107 pKa = 9.92SVYY110 pKa = 10.25ILGKK114 pKa = 9.79IGKK117 pKa = 8.34IWMDD121 pKa = 3.43EE122 pKa = 4.01NIKK125 pKa = 10.61LKK127 pKa = 10.73NHH129 pKa = 5.95TNSVMFWLVRR139 pKa = 11.84DD140 pKa = 3.7RR141 pKa = 11.84RR142 pKa = 11.84PYY144 pKa = 8.05GTPMDD149 pKa = 4.6FGQVFNMFDD158 pKa = 3.99NEE160 pKa = 4.11PSTATVKK167 pKa = 10.81NDD169 pKa = 2.84LRR171 pKa = 11.84DD172 pKa = 3.6RR173 pKa = 11.84YY174 pKa = 9.91QVMHH178 pKa = 7.14RR179 pKa = 11.84FHH181 pKa = 7.1AKK183 pKa = 8.01VTGGQYY189 pKa = 11.07ASNEE193 pKa = 3.78QAIVRR198 pKa = 11.84RR199 pKa = 11.84FWKK202 pKa = 10.31VNNHH206 pKa = 4.48VVYY209 pKa = 10.67NHH211 pKa = 5.8QEE213 pKa = 3.47AGKK216 pKa = 10.41YY217 pKa = 8.39EE218 pKa = 4.09NHH220 pKa = 6.52TEE222 pKa = 3.99NALLLYY228 pKa = 7.29MACTHH233 pKa = 7.07ASNPVYY239 pKa = 9.86ATLKK243 pKa = 9.47IRR245 pKa = 11.84IYY247 pKa = 10.66FYY249 pKa = 11.18DD250 pKa = 3.06SVMNN254 pKa = 4.52
Molecular weight: 29.73 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
969 |
87 |
368 |
193.8 |
22.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.47 ± 0.868 | 1.858 ± 0.378 |
4.85 ± 0.699 | 5.47 ± 0.863 |
3.715 ± 0.45 | 6.089 ± 0.793 |
4.231 ± 0.496 | 5.366 ± 0.995 |
5.057 ± 1.18 | 6.398 ± 0.961 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.167 ± 0.593 | 5.573 ± 0.929 |
5.986 ± 0.837 | 4.231 ± 0.833 |
8.05 ± 1.657 | 7.946 ± 1.739 |
5.366 ± 1.434 | 6.089 ± 1.193 |
1.858 ± 0.1 | 4.231 ± 1.05 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
