
Dentipellis fragilis
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetes incertae sedis; Russulales; Hericiaceae; Dentipellis
Average proteome isoelectric point is 6.82
Get precalculated fractions of proteins
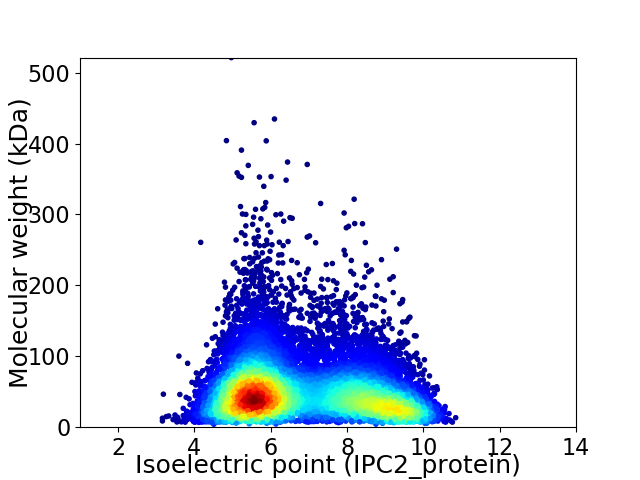
Virtual 2D-PAGE plot for 11715 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Y9Z0W0|A0A4Y9Z0W0_9AGAM Acetate--CoA ligase OS=Dentipellis fragilis OX=205917 GN=EVG20_g3933 PE=3 SV=1
MM1 pKa = 7.34FATLFTVTLLSAGAIKK17 pKa = 10.46GALAGLAINSPTLVQCEE34 pKa = 4.09DD35 pKa = 3.6AHH37 pKa = 6.88VSWASTKK44 pKa = 10.43GPYY47 pKa = 10.08NLIVTPGDD55 pKa = 3.95EE56 pKa = 4.39PCGDD60 pKa = 4.68AIVDD64 pKa = 4.35LGDD67 pKa = 3.93HH68 pKa = 7.39DD69 pKa = 4.75GTTMTYY75 pKa = 10.61SVALPAGQKK84 pKa = 10.12VLLSLQDD91 pKa = 3.73ANGDD95 pKa = 3.93EE96 pKa = 4.53AWSQEE101 pKa = 3.91LTVQAGSNTSCLPALLQVSSSISTSSVVPTVSSAKK136 pKa = 9.53FALSSTVVPLTAVTPATTLSAFALPSSDD164 pKa = 3.42PAAAAPAPSDD174 pKa = 3.54TVAGDD179 pKa = 4.38DD180 pKa = 4.41SDD182 pKa = 4.59SDD184 pKa = 4.01SPSVVGGAANAASNPFSGALAMHH207 pKa = 6.27QLYY210 pKa = 8.27TPAMLLSALATAFFVALL227 pKa = 4.01
MM1 pKa = 7.34FATLFTVTLLSAGAIKK17 pKa = 10.46GALAGLAINSPTLVQCEE34 pKa = 4.09DD35 pKa = 3.6AHH37 pKa = 6.88VSWASTKK44 pKa = 10.43GPYY47 pKa = 10.08NLIVTPGDD55 pKa = 3.95EE56 pKa = 4.39PCGDD60 pKa = 4.68AIVDD64 pKa = 4.35LGDD67 pKa = 3.93HH68 pKa = 7.39DD69 pKa = 4.75GTTMTYY75 pKa = 10.61SVALPAGQKK84 pKa = 10.12VLLSLQDD91 pKa = 3.73ANGDD95 pKa = 3.93EE96 pKa = 4.53AWSQEE101 pKa = 3.91LTVQAGSNTSCLPALLQVSSSISTSSVVPTVSSAKK136 pKa = 9.53FALSSTVVPLTAVTPATTLSAFALPSSDD164 pKa = 3.42PAAAAPAPSDD174 pKa = 3.54TVAGDD179 pKa = 4.38DD180 pKa = 4.41SDD182 pKa = 4.59SDD184 pKa = 4.01SPSVVGGAANAASNPFSGALAMHH207 pKa = 6.27QLYY210 pKa = 8.27TPAMLLSALATAFFVALL227 pKa = 4.01
Molecular weight: 22.62 kDa
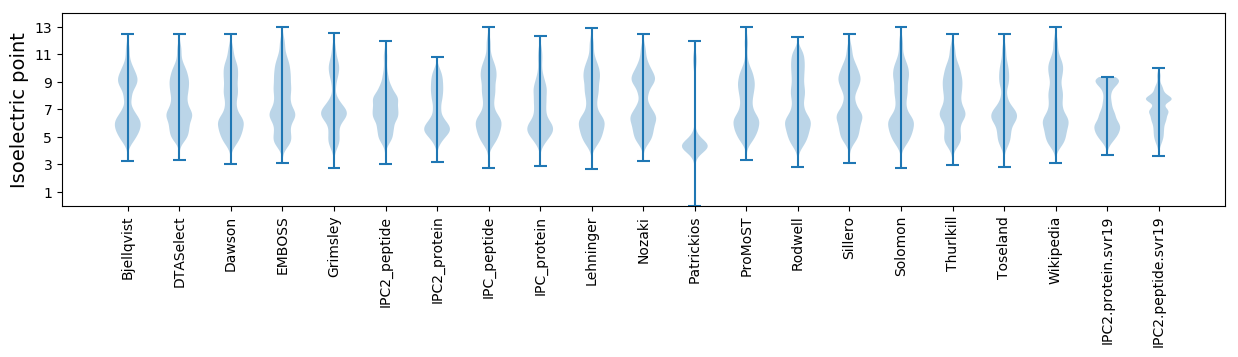
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Y9XNJ3|A0A4Y9XNJ3_9AGAM Uncharacterized protein OS=Dentipellis fragilis OX=205917 GN=EVG20_g10886 PE=4 SV=1
MM1 pKa = 7.84PLQAPMRR8 pKa = 11.84PSDD11 pKa = 3.75AVSPAAAHH19 pKa = 6.79RR20 pKa = 11.84KK21 pKa = 6.57LTPAARR27 pKa = 11.84PNSFHH32 pKa = 6.87NKK34 pKa = 6.68QHH36 pKa = 5.06TTNIPRR42 pKa = 11.84TQTRR46 pKa = 11.84SRR48 pKa = 11.84RR49 pKa = 11.84WQQNWNGTTTKK60 pKa = 10.31NDD62 pKa = 3.27RR63 pKa = 11.84AAGCTGLDD71 pKa = 3.32ARR73 pKa = 11.84HH74 pKa = 5.77VLWSLLSTNVMAGAVDD90 pKa = 3.44PHH92 pKa = 6.53TYY94 pKa = 9.5ICAGQPCGACVKK106 pKa = 10.26DD107 pKa = 3.4LRR109 pKa = 11.84GRR111 pKa = 11.84NVGEE115 pKa = 3.89KK116 pKa = 10.29RR117 pKa = 11.84EE118 pKa = 4.28ACVGRR123 pKa = 11.84AGPSEE128 pKa = 4.1ISQVIPIPKK137 pKa = 9.27ISIMQGARR145 pKa = 11.84ARR147 pKa = 11.84AFAFPDD153 pKa = 3.05KK154 pKa = 10.08KK155 pKa = 10.97ARR157 pKa = 11.84IISYY161 pKa = 6.83TSRR164 pKa = 11.84RR165 pKa = 11.84HH166 pKa = 5.04SAPPHH171 pKa = 6.11DD172 pKa = 4.16TLISNPDD179 pKa = 3.55TGTMTEE185 pKa = 4.54DD186 pKa = 2.98EE187 pKa = 4.72RR188 pKa = 11.84EE189 pKa = 4.19RR190 pKa = 11.84PTATAEE196 pKa = 3.97NDD198 pKa = 3.0NDD200 pKa = 3.53IANVWRR206 pKa = 11.84APTPYY211 pKa = 10.47ASSRR215 pKa = 11.84SDD217 pKa = 3.09AFAPDD222 pKa = 3.48TQEE225 pKa = 3.46QQQRR229 pKa = 11.84HH230 pKa = 4.64QHH232 pKa = 5.3HH233 pKa = 6.62CHH235 pKa = 5.72YY236 pKa = 10.49APHH239 pKa = 7.2ALRR242 pKa = 11.84GANSLATHH250 pKa = 7.44LYY252 pKa = 6.11PTSGTPRR259 pKa = 11.84APPPQAQTPRR269 pKa = 11.84AARR272 pKa = 11.84GAAPRR277 pKa = 11.84PPPRR281 pKa = 11.84GPSRR285 pKa = 11.84PPRR288 pKa = 11.84TSGARR293 pKa = 11.84TRR295 pKa = 11.84QTGASQLGMVTVTVTGVEE313 pKa = 4.2CWSRR317 pKa = 11.84SCNVNGDD324 pKa = 3.53RR325 pKa = 11.84EE326 pKa = 4.07RR327 pKa = 11.84DD328 pKa = 3.22VDD330 pKa = 4.2SFSLVSFRR338 pKa = 11.84QLAAPAEE345 pKa = 4.24MGMEE349 pKa = 3.62IAEE352 pKa = 4.15FAFTFKK358 pKa = 9.97FTSRR362 pKa = 11.84GLGDD366 pKa = 4.15PGAKK370 pKa = 9.22RR371 pKa = 11.84CIRR374 pKa = 11.84SRR376 pKa = 11.84SRR378 pKa = 11.84TEE380 pKa = 3.53TMRR383 pKa = 11.84ISILRR388 pKa = 11.84SDD390 pKa = 3.98LASYY394 pKa = 10.12CRR396 pKa = 11.84NNANMQTSTTYY407 pKa = 10.7HH408 pKa = 5.4EE409 pKa = 5.32AKK411 pKa = 10.34RR412 pKa = 11.84SMCLCLCPATQPGTTQTTYY431 pKa = 11.67ALRR434 pKa = 11.84TRR436 pKa = 11.84RR437 pKa = 11.84RR438 pKa = 11.84GIRR441 pKa = 11.84SDD443 pKa = 3.03KK444 pKa = 10.76HH445 pKa = 4.83KK446 pKa = 11.03APAKK450 pKa = 9.77RR451 pKa = 11.84APEE454 pKa = 3.71PHH456 pKa = 7.08LLRR459 pKa = 11.84EE460 pKa = 3.97QADD463 pKa = 3.41IEE465 pKa = 4.46VADD468 pKa = 4.06RR469 pKa = 11.84RR470 pKa = 11.84RR471 pKa = 11.84GRR473 pKa = 11.84EE474 pKa = 3.36HH475 pKa = 7.02RR476 pKa = 11.84LAVEE480 pKa = 3.95ARR482 pKa = 11.84RR483 pKa = 11.84LRR485 pKa = 11.84EE486 pKa = 3.97EE487 pKa = 4.04QEE489 pKa = 4.36HH490 pKa = 5.64VSARR494 pKa = 11.84HH495 pKa = 6.55DD496 pKa = 3.99GSQHH500 pKa = 5.79SISRR504 pKa = 11.84VASRR508 pKa = 11.84EE509 pKa = 3.84LGHH512 pKa = 6.25WVSHH516 pKa = 6.55IGRR519 pKa = 11.84TRR521 pKa = 11.84RR522 pKa = 11.84RR523 pKa = 11.84KK524 pKa = 9.94RR525 pKa = 11.84EE526 pKa = 3.54EE527 pKa = 3.57AKK529 pKa = 10.38IGEE532 pKa = 4.26KK533 pKa = 9.24TVYY536 pKa = 10.52SKK538 pKa = 7.4THH540 pKa = 6.23RR541 pKa = 11.84CGSDD545 pKa = 2.97PAXGVGCKK553 pKa = 10.37NSDD556 pKa = 3.55SAA558 pKa = 5.01
MM1 pKa = 7.84PLQAPMRR8 pKa = 11.84PSDD11 pKa = 3.75AVSPAAAHH19 pKa = 6.79RR20 pKa = 11.84KK21 pKa = 6.57LTPAARR27 pKa = 11.84PNSFHH32 pKa = 6.87NKK34 pKa = 6.68QHH36 pKa = 5.06TTNIPRR42 pKa = 11.84TQTRR46 pKa = 11.84SRR48 pKa = 11.84RR49 pKa = 11.84WQQNWNGTTTKK60 pKa = 10.31NDD62 pKa = 3.27RR63 pKa = 11.84AAGCTGLDD71 pKa = 3.32ARR73 pKa = 11.84HH74 pKa = 5.77VLWSLLSTNVMAGAVDD90 pKa = 3.44PHH92 pKa = 6.53TYY94 pKa = 9.5ICAGQPCGACVKK106 pKa = 10.26DD107 pKa = 3.4LRR109 pKa = 11.84GRR111 pKa = 11.84NVGEE115 pKa = 3.89KK116 pKa = 10.29RR117 pKa = 11.84EE118 pKa = 4.28ACVGRR123 pKa = 11.84AGPSEE128 pKa = 4.1ISQVIPIPKK137 pKa = 9.27ISIMQGARR145 pKa = 11.84ARR147 pKa = 11.84AFAFPDD153 pKa = 3.05KK154 pKa = 10.08KK155 pKa = 10.97ARR157 pKa = 11.84IISYY161 pKa = 6.83TSRR164 pKa = 11.84RR165 pKa = 11.84HH166 pKa = 5.04SAPPHH171 pKa = 6.11DD172 pKa = 4.16TLISNPDD179 pKa = 3.55TGTMTEE185 pKa = 4.54DD186 pKa = 2.98EE187 pKa = 4.72RR188 pKa = 11.84EE189 pKa = 4.19RR190 pKa = 11.84PTATAEE196 pKa = 3.97NDD198 pKa = 3.0NDD200 pKa = 3.53IANVWRR206 pKa = 11.84APTPYY211 pKa = 10.47ASSRR215 pKa = 11.84SDD217 pKa = 3.09AFAPDD222 pKa = 3.48TQEE225 pKa = 3.46QQQRR229 pKa = 11.84HH230 pKa = 4.64QHH232 pKa = 5.3HH233 pKa = 6.62CHH235 pKa = 5.72YY236 pKa = 10.49APHH239 pKa = 7.2ALRR242 pKa = 11.84GANSLATHH250 pKa = 7.44LYY252 pKa = 6.11PTSGTPRR259 pKa = 11.84APPPQAQTPRR269 pKa = 11.84AARR272 pKa = 11.84GAAPRR277 pKa = 11.84PPPRR281 pKa = 11.84GPSRR285 pKa = 11.84PPRR288 pKa = 11.84TSGARR293 pKa = 11.84TRR295 pKa = 11.84QTGASQLGMVTVTVTGVEE313 pKa = 4.2CWSRR317 pKa = 11.84SCNVNGDD324 pKa = 3.53RR325 pKa = 11.84EE326 pKa = 4.07RR327 pKa = 11.84DD328 pKa = 3.22VDD330 pKa = 4.2SFSLVSFRR338 pKa = 11.84QLAAPAEE345 pKa = 4.24MGMEE349 pKa = 3.62IAEE352 pKa = 4.15FAFTFKK358 pKa = 9.97FTSRR362 pKa = 11.84GLGDD366 pKa = 4.15PGAKK370 pKa = 9.22RR371 pKa = 11.84CIRR374 pKa = 11.84SRR376 pKa = 11.84SRR378 pKa = 11.84TEE380 pKa = 3.53TMRR383 pKa = 11.84ISILRR388 pKa = 11.84SDD390 pKa = 3.98LASYY394 pKa = 10.12CRR396 pKa = 11.84NNANMQTSTTYY407 pKa = 10.7HH408 pKa = 5.4EE409 pKa = 5.32AKK411 pKa = 10.34RR412 pKa = 11.84SMCLCLCPATQPGTTQTTYY431 pKa = 11.67ALRR434 pKa = 11.84TRR436 pKa = 11.84RR437 pKa = 11.84RR438 pKa = 11.84GIRR441 pKa = 11.84SDD443 pKa = 3.03KK444 pKa = 10.76HH445 pKa = 4.83KK446 pKa = 11.03APAKK450 pKa = 9.77RR451 pKa = 11.84APEE454 pKa = 3.71PHH456 pKa = 7.08LLRR459 pKa = 11.84EE460 pKa = 3.97QADD463 pKa = 3.41IEE465 pKa = 4.46VADD468 pKa = 4.06RR469 pKa = 11.84RR470 pKa = 11.84RR471 pKa = 11.84GRR473 pKa = 11.84EE474 pKa = 3.36HH475 pKa = 7.02RR476 pKa = 11.84LAVEE480 pKa = 3.95ARR482 pKa = 11.84RR483 pKa = 11.84LRR485 pKa = 11.84EE486 pKa = 3.97EE487 pKa = 4.04QEE489 pKa = 4.36HH490 pKa = 5.64VSARR494 pKa = 11.84HH495 pKa = 6.55DD496 pKa = 3.99GSQHH500 pKa = 5.79SISRR504 pKa = 11.84VASRR508 pKa = 11.84EE509 pKa = 3.84LGHH512 pKa = 6.25WVSHH516 pKa = 6.55IGRR519 pKa = 11.84TRR521 pKa = 11.84RR522 pKa = 11.84RR523 pKa = 11.84KK524 pKa = 9.94RR525 pKa = 11.84EE526 pKa = 3.54EE527 pKa = 3.57AKK529 pKa = 10.38IGEE532 pKa = 4.26KK533 pKa = 9.24TVYY536 pKa = 10.52SKK538 pKa = 7.4THH540 pKa = 6.23RR541 pKa = 11.84CGSDD545 pKa = 2.97PAXGVGCKK553 pKa = 10.37NSDD556 pKa = 3.55SAA558 pKa = 5.01
Molecular weight: 61.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5974488 |
38 |
4685 |
510.0 |
56.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.399 ± 0.022 | 1.365 ± 0.009 |
5.552 ± 0.014 | 5.635 ± 0.02 |
3.552 ± 0.014 | 6.408 ± 0.017 |
2.773 ± 0.01 | 4.507 ± 0.017 |
4.099 ± 0.017 | 9.207 ± 0.027 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.042 ± 0.008 | 3.063 ± 0.012 |
7.01 ± 0.027 | 3.628 ± 0.014 |
6.771 ± 0.02 | 8.674 ± 0.026 |
5.965 ± 0.013 | 6.312 ± 0.014 |
1.397 ± 0.009 | 2.533 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
