
Newcastle disease virus (strain Chicken/United States/B1/48) (NDV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Avulavirinae; Orthoavulavirus; Avian orthoavulavirus 1
Average proteome isoelectric point is 6.83
Get precalculated fractions of proteins
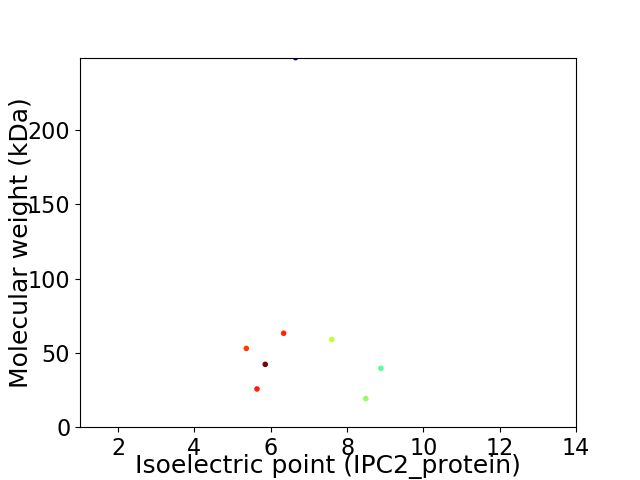
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
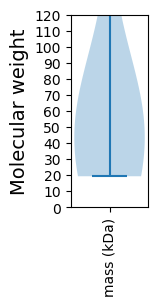
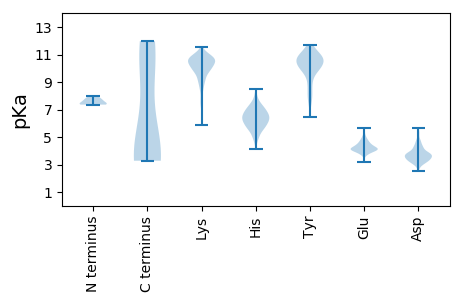
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q91UL0|HN_NDVB1 Hemagglutinin-neuraminidase OS=Newcastle disease virus (strain Chicken/United States/B1/48) OX=652953 GN=HN PE=1 SV=1
MM1 pKa = 7.45SSVFDD6 pKa = 4.24EE7 pKa = 4.58YY8 pKa = 11.32EE9 pKa = 3.81QLLAAQTRR17 pKa = 11.84PNGAHH22 pKa = 6.5GGGEE26 pKa = 3.98KK27 pKa = 10.82GSTLKK32 pKa = 10.74VDD34 pKa = 3.72VPVFTLNSDD43 pKa = 3.68DD44 pKa = 5.54PEE46 pKa = 4.53DD47 pKa = 3.22RR48 pKa = 11.84WSFVVFCLRR57 pKa = 11.84IAVSEE62 pKa = 4.28DD63 pKa = 3.01ANKK66 pKa = 10.05PLRR69 pKa = 11.84QGALISLLCSHH80 pKa = 6.17SQVMRR85 pKa = 11.84NHH87 pKa = 5.78VALAGKK93 pKa = 9.58QNEE96 pKa = 4.04ATLAVLEE103 pKa = 4.36IDD105 pKa = 4.13GFANGTPQFNNRR117 pKa = 11.84SGVSEE122 pKa = 3.88EE123 pKa = 3.83RR124 pKa = 11.84AQRR127 pKa = 11.84FAMIAGSLPRR137 pKa = 11.84ACSNGTPFVTAGAEE151 pKa = 3.83DD152 pKa = 4.74DD153 pKa = 4.08APEE156 pKa = 5.68DD157 pKa = 3.31ITDD160 pKa = 3.59TLEE163 pKa = 4.86RR164 pKa = 11.84ILSIQAQVWVTVAKK178 pKa = 10.9AMTAYY183 pKa = 7.35EE184 pKa = 4.12TADD187 pKa = 3.26EE188 pKa = 4.57SEE190 pKa = 3.98TRR192 pKa = 11.84RR193 pKa = 11.84INKK196 pKa = 9.01YY197 pKa = 6.52MQQGRR202 pKa = 11.84VQKK205 pKa = 10.55KK206 pKa = 7.89YY207 pKa = 10.65ILYY210 pKa = 7.91PVCRR214 pKa = 11.84STIQLTIRR222 pKa = 11.84QSLAVRR228 pKa = 11.84IFLVSEE234 pKa = 4.38LKK236 pKa = 10.46RR237 pKa = 11.84GRR239 pKa = 11.84NTAGGTSTYY248 pKa = 10.11YY249 pKa = 11.25NLVGDD254 pKa = 3.76VDD256 pKa = 4.37SYY258 pKa = 11.6IRR260 pKa = 11.84NTGLTAFFLTLKK272 pKa = 10.69YY273 pKa = 10.63GINTKK278 pKa = 9.69TSALALSSLSGDD290 pKa = 3.33IQKK293 pKa = 9.34MKK295 pKa = 10.85QLMRR299 pKa = 11.84LYY301 pKa = 10.82RR302 pKa = 11.84MKK304 pKa = 10.88GDD306 pKa = 3.4NAPYY310 pKa = 7.51MTLLGDD316 pKa = 3.71SDD318 pKa = 3.87QMSFAPAEE326 pKa = 4.05YY327 pKa = 9.98AQLYY331 pKa = 8.71SFAMGMASVLDD342 pKa = 4.0KK343 pKa = 11.15GTGKK347 pKa = 10.26YY348 pKa = 8.36QFAKK352 pKa = 10.8DD353 pKa = 3.85FMSTSFWRR361 pKa = 11.84LGVEE365 pKa = 4.05YY366 pKa = 10.28AQAQGSSINEE376 pKa = 3.69DD377 pKa = 2.85MAAEE381 pKa = 4.45LKK383 pKa = 8.96LTPAARR389 pKa = 11.84RR390 pKa = 11.84GLAAAAQRR398 pKa = 11.84VSEE401 pKa = 4.41VTSSIDD407 pKa = 3.23MPTQQVGVLTGLSEE421 pKa = 4.57GGSQALQGGSNRR433 pKa = 11.84SQGQPEE439 pKa = 4.24AGDD442 pKa = 4.17GEE444 pKa = 4.82TQFLDD449 pKa = 4.29LMRR452 pKa = 11.84AVANSMRR459 pKa = 11.84EE460 pKa = 4.25APNSAQGTPQSGPPPTPGPSQDD482 pKa = 4.47NDD484 pKa = 3.98TDD486 pKa = 2.94WGYY489 pKa = 11.97
MM1 pKa = 7.45SSVFDD6 pKa = 4.24EE7 pKa = 4.58YY8 pKa = 11.32EE9 pKa = 3.81QLLAAQTRR17 pKa = 11.84PNGAHH22 pKa = 6.5GGGEE26 pKa = 3.98KK27 pKa = 10.82GSTLKK32 pKa = 10.74VDD34 pKa = 3.72VPVFTLNSDD43 pKa = 3.68DD44 pKa = 5.54PEE46 pKa = 4.53DD47 pKa = 3.22RR48 pKa = 11.84WSFVVFCLRR57 pKa = 11.84IAVSEE62 pKa = 4.28DD63 pKa = 3.01ANKK66 pKa = 10.05PLRR69 pKa = 11.84QGALISLLCSHH80 pKa = 6.17SQVMRR85 pKa = 11.84NHH87 pKa = 5.78VALAGKK93 pKa = 9.58QNEE96 pKa = 4.04ATLAVLEE103 pKa = 4.36IDD105 pKa = 4.13GFANGTPQFNNRR117 pKa = 11.84SGVSEE122 pKa = 3.88EE123 pKa = 3.83RR124 pKa = 11.84AQRR127 pKa = 11.84FAMIAGSLPRR137 pKa = 11.84ACSNGTPFVTAGAEE151 pKa = 3.83DD152 pKa = 4.74DD153 pKa = 4.08APEE156 pKa = 5.68DD157 pKa = 3.31ITDD160 pKa = 3.59TLEE163 pKa = 4.86RR164 pKa = 11.84ILSIQAQVWVTVAKK178 pKa = 10.9AMTAYY183 pKa = 7.35EE184 pKa = 4.12TADD187 pKa = 3.26EE188 pKa = 4.57SEE190 pKa = 3.98TRR192 pKa = 11.84RR193 pKa = 11.84INKK196 pKa = 9.01YY197 pKa = 6.52MQQGRR202 pKa = 11.84VQKK205 pKa = 10.55KK206 pKa = 7.89YY207 pKa = 10.65ILYY210 pKa = 7.91PVCRR214 pKa = 11.84STIQLTIRR222 pKa = 11.84QSLAVRR228 pKa = 11.84IFLVSEE234 pKa = 4.38LKK236 pKa = 10.46RR237 pKa = 11.84GRR239 pKa = 11.84NTAGGTSTYY248 pKa = 10.11YY249 pKa = 11.25NLVGDD254 pKa = 3.76VDD256 pKa = 4.37SYY258 pKa = 11.6IRR260 pKa = 11.84NTGLTAFFLTLKK272 pKa = 10.69YY273 pKa = 10.63GINTKK278 pKa = 9.69TSALALSSLSGDD290 pKa = 3.33IQKK293 pKa = 9.34MKK295 pKa = 10.85QLMRR299 pKa = 11.84LYY301 pKa = 10.82RR302 pKa = 11.84MKK304 pKa = 10.88GDD306 pKa = 3.4NAPYY310 pKa = 7.51MTLLGDD316 pKa = 3.71SDD318 pKa = 3.87QMSFAPAEE326 pKa = 4.05YY327 pKa = 9.98AQLYY331 pKa = 8.71SFAMGMASVLDD342 pKa = 4.0KK343 pKa = 11.15GTGKK347 pKa = 10.26YY348 pKa = 8.36QFAKK352 pKa = 10.8DD353 pKa = 3.85FMSTSFWRR361 pKa = 11.84LGVEE365 pKa = 4.05YY366 pKa = 10.28AQAQGSSINEE376 pKa = 3.69DD377 pKa = 2.85MAAEE381 pKa = 4.45LKK383 pKa = 8.96LTPAARR389 pKa = 11.84RR390 pKa = 11.84GLAAAAQRR398 pKa = 11.84VSEE401 pKa = 4.41VTSSIDD407 pKa = 3.23MPTQQVGVLTGLSEE421 pKa = 4.57GGSQALQGGSNRR433 pKa = 11.84SQGQPEE439 pKa = 4.24AGDD442 pKa = 4.17GEE444 pKa = 4.82TQFLDD449 pKa = 4.29LMRR452 pKa = 11.84AVANSMRR459 pKa = 11.84EE460 pKa = 4.25APNSAQGTPQSGPPPTPGPSQDD482 pKa = 4.47NDD484 pKa = 3.98TDD486 pKa = 2.94WGYY489 pKa = 11.97
Molecular weight: 53.02 kDa
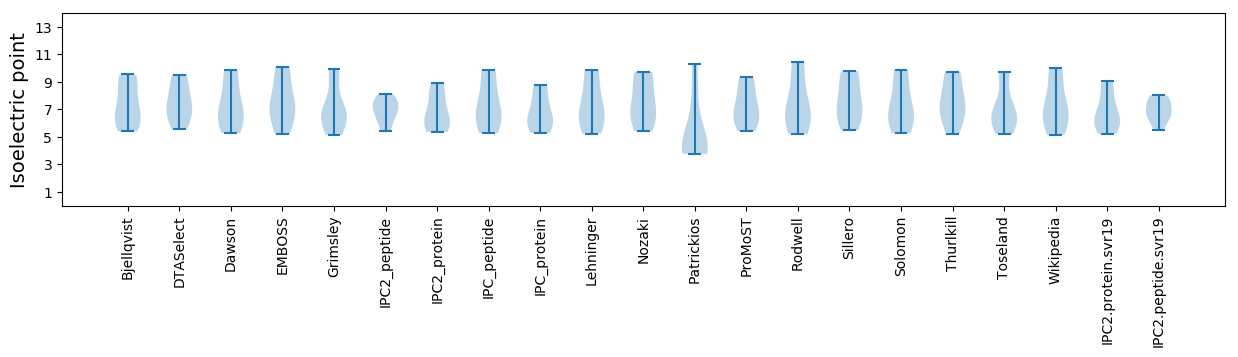
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q9DLD6|PHOSP_NDVB1 Phosphoprotein OS=Newcastle disease virus (strain Chicken/United States/B1/48) OX=652953 GN=P PE=3 SV=1
MM1 pKa = 7.99DD2 pKa = 4.33SSRR5 pKa = 11.84TIGLYY10 pKa = 10.09FDD12 pKa = 4.63SAHH15 pKa = 6.53SSSNLLAFPIVLQDD29 pKa = 3.17TGDD32 pKa = 3.69GKK34 pKa = 11.01KK35 pKa = 10.16QIAPQYY41 pKa = 9.71RR42 pKa = 11.84IQRR45 pKa = 11.84LDD47 pKa = 2.92LWTDD51 pKa = 3.2SKK53 pKa = 11.18EE54 pKa = 3.97DD55 pKa = 3.45SVFITTYY62 pKa = 10.97GFIFQVGNEE71 pKa = 3.98EE72 pKa = 4.1ATVGIIDD79 pKa = 4.73DD80 pKa = 3.78KK81 pKa = 11.11PKK83 pKa = 10.89RR84 pKa = 11.84EE85 pKa = 4.12LLSAAMLCLGSVPNTGDD102 pKa = 3.72LIEE105 pKa = 4.64LARR108 pKa = 11.84ACLTMMVTCKK118 pKa = 10.27KK119 pKa = 10.43SATNTEE125 pKa = 3.74RR126 pKa = 11.84MVFSVVQAPQVLQSCRR142 pKa = 11.84VVANKK147 pKa = 10.08YY148 pKa = 10.08SSVNAVKK155 pKa = 10.22HH156 pKa = 4.84VKK158 pKa = 10.26APEE161 pKa = 4.53KK162 pKa = 10.52IPGSGTLEE170 pKa = 3.93YY171 pKa = 10.56KK172 pKa = 10.96VNFVSLTVVPKK183 pKa = 10.37KK184 pKa = 10.45DD185 pKa = 3.43VYY187 pKa = 10.74KK188 pKa = 10.75IPAAVLKK195 pKa = 10.62ISGSSLYY202 pKa = 10.87NLALNVTINVEE213 pKa = 3.83VDD215 pKa = 3.04PRR217 pKa = 11.84SPLVKK222 pKa = 10.22SLSKK226 pKa = 10.64SDD228 pKa = 3.09SGYY231 pKa = 9.64YY232 pKa = 10.76ANLFLHH238 pKa = 6.98IGLMTTVDD246 pKa = 3.37RR247 pKa = 11.84KK248 pKa = 10.11GKK250 pKa = 10.33KK251 pKa = 7.01VTFDD255 pKa = 3.32KK256 pKa = 11.21LEE258 pKa = 4.05KK259 pKa = 10.22KK260 pKa = 9.79IRR262 pKa = 11.84SLDD265 pKa = 3.67LSVGLSDD272 pKa = 3.83VLGPSVLVKK281 pKa = 10.85ARR283 pKa = 11.84GARR286 pKa = 11.84TKK288 pKa = 10.73LLAPFFSSSGTACYY302 pKa = 9.57PIANASPQVAKK313 pKa = 10.63ILWSQTACLRR323 pKa = 11.84SVKK326 pKa = 10.15IIIQAGTQRR335 pKa = 11.84AVAVTADD342 pKa = 3.79HH343 pKa = 6.67EE344 pKa = 4.77VTSTKK349 pKa = 10.49LEE351 pKa = 4.33KK352 pKa = 10.74GHH354 pKa = 6.49TLAKK358 pKa = 10.37YY359 pKa = 10.7NPFKK363 pKa = 10.84KK364 pKa = 10.49
MM1 pKa = 7.99DD2 pKa = 4.33SSRR5 pKa = 11.84TIGLYY10 pKa = 10.09FDD12 pKa = 4.63SAHH15 pKa = 6.53SSSNLLAFPIVLQDD29 pKa = 3.17TGDD32 pKa = 3.69GKK34 pKa = 11.01KK35 pKa = 10.16QIAPQYY41 pKa = 9.71RR42 pKa = 11.84IQRR45 pKa = 11.84LDD47 pKa = 2.92LWTDD51 pKa = 3.2SKK53 pKa = 11.18EE54 pKa = 3.97DD55 pKa = 3.45SVFITTYY62 pKa = 10.97GFIFQVGNEE71 pKa = 3.98EE72 pKa = 4.1ATVGIIDD79 pKa = 4.73DD80 pKa = 3.78KK81 pKa = 11.11PKK83 pKa = 10.89RR84 pKa = 11.84EE85 pKa = 4.12LLSAAMLCLGSVPNTGDD102 pKa = 3.72LIEE105 pKa = 4.64LARR108 pKa = 11.84ACLTMMVTCKK118 pKa = 10.27KK119 pKa = 10.43SATNTEE125 pKa = 3.74RR126 pKa = 11.84MVFSVVQAPQVLQSCRR142 pKa = 11.84VVANKK147 pKa = 10.08YY148 pKa = 10.08SSVNAVKK155 pKa = 10.22HH156 pKa = 4.84VKK158 pKa = 10.26APEE161 pKa = 4.53KK162 pKa = 10.52IPGSGTLEE170 pKa = 3.93YY171 pKa = 10.56KK172 pKa = 10.96VNFVSLTVVPKK183 pKa = 10.37KK184 pKa = 10.45DD185 pKa = 3.43VYY187 pKa = 10.74KK188 pKa = 10.75IPAAVLKK195 pKa = 10.62ISGSSLYY202 pKa = 10.87NLALNVTINVEE213 pKa = 3.83VDD215 pKa = 3.04PRR217 pKa = 11.84SPLVKK222 pKa = 10.22SLSKK226 pKa = 10.64SDD228 pKa = 3.09SGYY231 pKa = 9.64YY232 pKa = 10.76ANLFLHH238 pKa = 6.98IGLMTTVDD246 pKa = 3.37RR247 pKa = 11.84KK248 pKa = 10.11GKK250 pKa = 10.33KK251 pKa = 7.01VTFDD255 pKa = 3.32KK256 pKa = 11.21LEE258 pKa = 4.05KK259 pKa = 10.22KK260 pKa = 9.79IRR262 pKa = 11.84SLDD265 pKa = 3.67LSVGLSDD272 pKa = 3.83VLGPSVLVKK281 pKa = 10.85ARR283 pKa = 11.84GARR286 pKa = 11.84TKK288 pKa = 10.73LLAPFFSSSGTACYY302 pKa = 9.57PIANASPQVAKK313 pKa = 10.63ILWSQTACLRR323 pKa = 11.84SVKK326 pKa = 10.15IIIQAGTQRR335 pKa = 11.84AVAVTADD342 pKa = 3.79HH343 pKa = 6.67EE344 pKa = 4.77VTSTKK349 pKa = 10.49LEE351 pKa = 4.33KK352 pKa = 10.74GHH354 pKa = 6.49TLAKK358 pKa = 10.37YY359 pKa = 10.7NPFKK363 pKa = 10.84KK364 pKa = 10.49
Molecular weight: 39.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5000 |
179 |
2204 |
625.0 |
68.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.46 ± 0.67 | 1.68 ± 0.218 |
5.16 ± 0.276 | 4.82 ± 0.402 |
3.2 ± 0.483 | 6.04 ± 0.755 |
2.02 ± 0.488 | 6.0 ± 0.571 |
5.14 ± 0.489 | 10.04 ± 1.077 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.5 ± 0.215 | 4.64 ± 0.344 |
5.16 ± 0.663 | 4.34 ± 0.759 |
5.04 ± 0.466 | 9.2 ± 0.476 |
7.36 ± 0.687 | 6.28 ± 0.392 |
0.84 ± 0.133 | 3.08 ± 0.383 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
