
bacterium HR15
Taxonomy: cellular organisms; Bacteria; unclassified Bacteria
Average proteome isoelectric point is 6.68
Get precalculated fractions of proteins
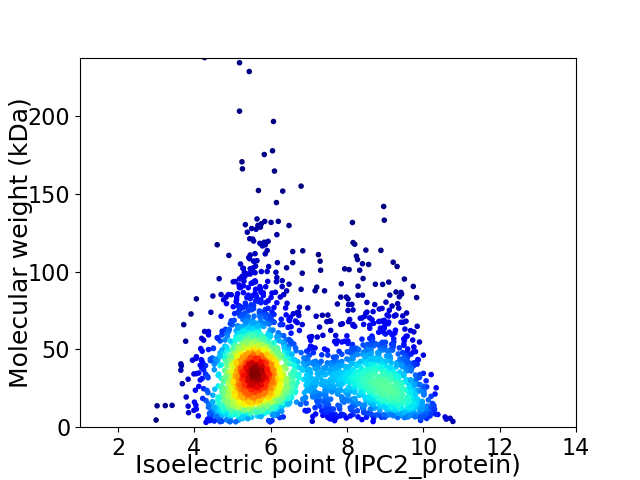
Virtual 2D-PAGE plot for 2671 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H5WS41|A0A2H5WS41_9BACT Site-specific DNA-methyltransferase (adenine-specific) OS=bacterium HR15 OX=2035410 GN=dpnM_1 PE=3 SV=1
MM1 pKa = 7.09MMNRR5 pKa = 11.84LIALPIALAFVCGLVLAQPRR25 pKa = 11.84IVINEE30 pKa = 4.1FAYY33 pKa = 10.7DD34 pKa = 3.72DD35 pKa = 3.92TSTDD39 pKa = 3.13DD40 pKa = 3.54VEE42 pKa = 5.47FVEE45 pKa = 6.06LYY47 pKa = 10.77NADD50 pKa = 4.03TVPVDD55 pKa = 3.08ISGWRR60 pKa = 11.84LEE62 pKa = 4.29SGDD65 pKa = 4.82LVSGDD70 pKa = 4.04NNPDD74 pKa = 3.24YY75 pKa = 10.02TIPGAPGSGTVVLQPGQFYY94 pKa = 10.98VIGSEE99 pKa = 4.21LVPNVNLVVGANNLFEE115 pKa = 5.86NDD117 pKa = 4.01NEE119 pKa = 4.08WTALKK124 pKa = 9.76MPDD127 pKa = 3.73GMVVDD132 pKa = 5.84AIAYY136 pKa = 5.35EE137 pKa = 4.49TNKK140 pKa = 10.52APDD143 pKa = 3.34LTTWVPANIIPQLGRR158 pKa = 11.84GIWGNYY164 pKa = 5.24ITLQASTTTDD174 pKa = 3.35DD175 pKa = 4.0TLLSLQRR182 pKa = 11.84WRR184 pKa = 11.84DD185 pKa = 3.59GLDD188 pKa = 3.21TNDD191 pKa = 3.24NGRR194 pKa = 11.84DD195 pKa = 3.8FGHH198 pKa = 6.33MPWTPGASNNTTPTPFSQNFNALNVYY224 pKa = 10.51DD225 pKa = 4.93LVPNLSGSFRR235 pKa = 11.84APRR238 pKa = 11.84VIDD241 pKa = 3.41PTQCDD246 pKa = 3.21TTYY249 pKa = 10.66ATTPNPNCIPASPDD263 pKa = 2.83GGNAMIGWDD272 pKa = 3.27WAGGGNMATSTFAFQNRR289 pKa = 11.84AGYY292 pKa = 10.34DD293 pKa = 3.32LLIYY297 pKa = 9.79IDD299 pKa = 4.15NSMPIPGEE307 pKa = 3.98LEE309 pKa = 3.52SWMIGLMGTCDD320 pKa = 3.35SFYY323 pKa = 10.67NIPFRR328 pKa = 11.84CLVNASGPTGVGWMFKK344 pKa = 10.22RR345 pKa = 11.84ANTANDD351 pKa = 3.23PMEE354 pKa = 4.43VKK356 pKa = 10.51LRR358 pKa = 11.84LVDD361 pKa = 4.33AGPGGDD367 pKa = 4.23SNPGCGTAGTMQWTTFGEE385 pKa = 4.23IDD387 pKa = 3.94LSNYY391 pKa = 7.85TPGWYY396 pKa = 9.9RR397 pKa = 11.84LRR399 pKa = 11.84LEE401 pKa = 4.22VKK403 pKa = 8.95GDD405 pKa = 3.53KK406 pKa = 10.8VVGTFSNPDD415 pKa = 2.82GSGAITFSGTTEE427 pKa = 3.94GCIGAFYY434 pKa = 10.24IGYY437 pKa = 9.35RR438 pKa = 11.84EE439 pKa = 4.93AYY441 pKa = 6.01TTDD444 pKa = 3.24PFANPVRR451 pKa = 11.84IDD453 pKa = 3.53ALSLFIPTDD462 pKa = 3.38GDD464 pKa = 3.45VDD466 pKa = 4.26GNGCVDD472 pKa = 4.28DD473 pKa = 5.59ADD475 pKa = 4.1LLEE478 pKa = 4.02VLFAFGATGCTRR490 pKa = 11.84ADD492 pKa = 3.47VNGDD496 pKa = 3.72GVVDD500 pKa = 4.42DD501 pKa = 5.53ADD503 pKa = 4.19LLTVLFDD510 pKa = 4.6FGSGCC515 pKa = 4.3
MM1 pKa = 7.09MMNRR5 pKa = 11.84LIALPIALAFVCGLVLAQPRR25 pKa = 11.84IVINEE30 pKa = 4.1FAYY33 pKa = 10.7DD34 pKa = 3.72DD35 pKa = 3.92TSTDD39 pKa = 3.13DD40 pKa = 3.54VEE42 pKa = 5.47FVEE45 pKa = 6.06LYY47 pKa = 10.77NADD50 pKa = 4.03TVPVDD55 pKa = 3.08ISGWRR60 pKa = 11.84LEE62 pKa = 4.29SGDD65 pKa = 4.82LVSGDD70 pKa = 4.04NNPDD74 pKa = 3.24YY75 pKa = 10.02TIPGAPGSGTVVLQPGQFYY94 pKa = 10.98VIGSEE99 pKa = 4.21LVPNVNLVVGANNLFEE115 pKa = 5.86NDD117 pKa = 4.01NEE119 pKa = 4.08WTALKK124 pKa = 9.76MPDD127 pKa = 3.73GMVVDD132 pKa = 5.84AIAYY136 pKa = 5.35EE137 pKa = 4.49TNKK140 pKa = 10.52APDD143 pKa = 3.34LTTWVPANIIPQLGRR158 pKa = 11.84GIWGNYY164 pKa = 5.24ITLQASTTTDD174 pKa = 3.35DD175 pKa = 4.0TLLSLQRR182 pKa = 11.84WRR184 pKa = 11.84DD185 pKa = 3.59GLDD188 pKa = 3.21TNDD191 pKa = 3.24NGRR194 pKa = 11.84DD195 pKa = 3.8FGHH198 pKa = 6.33MPWTPGASNNTTPTPFSQNFNALNVYY224 pKa = 10.51DD225 pKa = 4.93LVPNLSGSFRR235 pKa = 11.84APRR238 pKa = 11.84VIDD241 pKa = 3.41PTQCDD246 pKa = 3.21TTYY249 pKa = 10.66ATTPNPNCIPASPDD263 pKa = 2.83GGNAMIGWDD272 pKa = 3.27WAGGGNMATSTFAFQNRR289 pKa = 11.84AGYY292 pKa = 10.34DD293 pKa = 3.32LLIYY297 pKa = 9.79IDD299 pKa = 4.15NSMPIPGEE307 pKa = 3.98LEE309 pKa = 3.52SWMIGLMGTCDD320 pKa = 3.35SFYY323 pKa = 10.67NIPFRR328 pKa = 11.84CLVNASGPTGVGWMFKK344 pKa = 10.22RR345 pKa = 11.84ANTANDD351 pKa = 3.23PMEE354 pKa = 4.43VKK356 pKa = 10.51LRR358 pKa = 11.84LVDD361 pKa = 4.33AGPGGDD367 pKa = 4.23SNPGCGTAGTMQWTTFGEE385 pKa = 4.23IDD387 pKa = 3.94LSNYY391 pKa = 7.85TPGWYY396 pKa = 9.9RR397 pKa = 11.84LRR399 pKa = 11.84LEE401 pKa = 4.22VKK403 pKa = 8.95GDD405 pKa = 3.53KK406 pKa = 10.8VVGTFSNPDD415 pKa = 2.82GSGAITFSGTTEE427 pKa = 3.94GCIGAFYY434 pKa = 10.24IGYY437 pKa = 9.35RR438 pKa = 11.84EE439 pKa = 4.93AYY441 pKa = 6.01TTDD444 pKa = 3.24PFANPVRR451 pKa = 11.84IDD453 pKa = 3.53ALSLFIPTDD462 pKa = 3.38GDD464 pKa = 3.45VDD466 pKa = 4.26GNGCVDD472 pKa = 4.28DD473 pKa = 5.59ADD475 pKa = 4.1LLEE478 pKa = 4.02VLFAFGATGCTRR490 pKa = 11.84ADD492 pKa = 3.47VNGDD496 pKa = 3.72GVVDD500 pKa = 4.42DD501 pKa = 5.53ADD503 pKa = 4.19LLTVLFDD510 pKa = 4.6FGSGCC515 pKa = 4.3
Molecular weight: 55.27 kDa
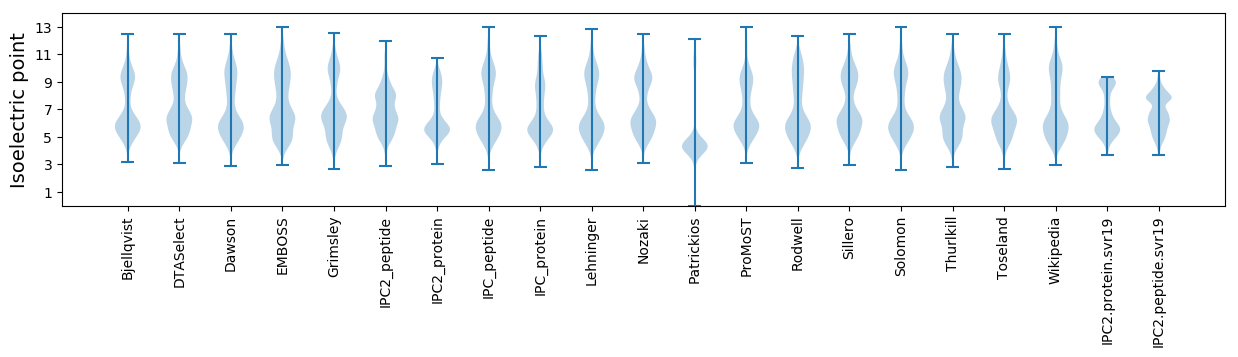
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H5WXW9|A0A2H5WXW9_9BACT Uncharacterized protein OS=bacterium HR15 OX=2035410 GN=HRbin15_02295 PE=4 SV=1
MM1 pKa = 6.96FRR3 pKa = 11.84KK4 pKa = 10.09VLGKK8 pKa = 10.06RR9 pKa = 11.84LKK11 pKa = 9.94TLTTRR16 pKa = 11.84FNSALQVVKK25 pKa = 10.56IRR27 pKa = 11.84LLSVVV32 pKa = 3.49
MM1 pKa = 6.96FRR3 pKa = 11.84KK4 pKa = 10.09VLGKK8 pKa = 10.06RR9 pKa = 11.84LKK11 pKa = 9.94TLTTRR16 pKa = 11.84FNSALQVVKK25 pKa = 10.56IRR27 pKa = 11.84LLSVVV32 pKa = 3.49
Molecular weight: 3.72 kDa
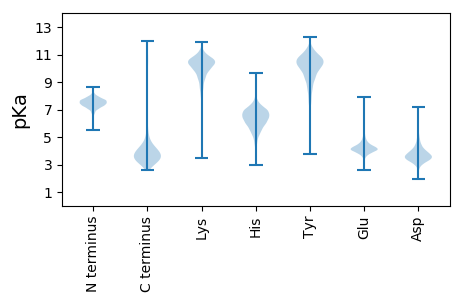
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
916987 |
29 |
2176 |
343.3 |
38.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.481 ± 0.051 | 1.06 ± 0.022 |
4.532 ± 0.038 | 6.605 ± 0.053 |
3.541 ± 0.029 | 7.716 ± 0.054 |
2.292 ± 0.027 | 5.201 ± 0.033 |
2.704 ± 0.031 | 11.207 ± 0.072 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.223 ± 0.022 | 2.824 ± 0.037 |
5.818 ± 0.036 | 4.625 ± 0.044 |
7.827 ± 0.043 | 4.835 ± 0.041 |
5.218 ± 0.038 | 7.229 ± 0.04 |
1.88 ± 0.027 | 3.182 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
