
Rhodobacteraceae bacterium SB2
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; unclassified Rhodobacteraceae
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
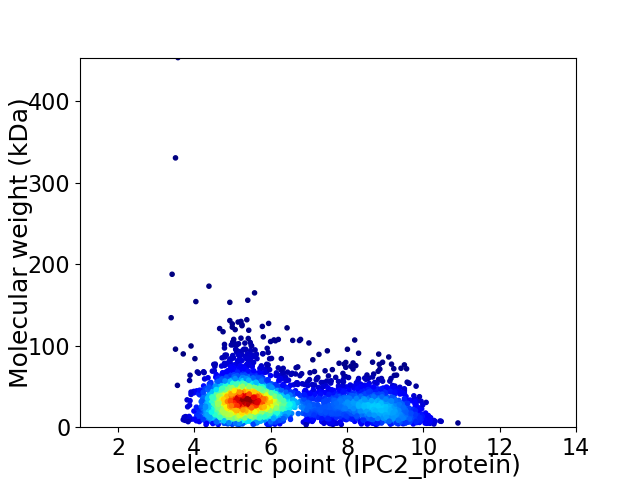
Virtual 2D-PAGE plot for 3489 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A177H6R6|A0A177H6R6_9RHOB Bacterial regulatory proteinC Fis family OS=Rhodobacteraceae bacterium SB2 OX=1689867 GN=pfor_25c2641 PE=4 SV=1
MM1 pKa = 7.77AIKK4 pKa = 10.37SVTVFIDD11 pKa = 3.24QFSDD15 pKa = 3.68RR16 pKa = 11.84IFDD19 pKa = 4.17LGHH22 pKa = 6.82DD23 pKa = 4.02NIYY26 pKa = 10.7LYY28 pKa = 10.49DD29 pKa = 3.67YY30 pKa = 11.33GNFHH34 pKa = 5.93EE35 pKa = 5.29TVWTDD40 pKa = 2.5NYY42 pKa = 10.85YY43 pKa = 10.93NVSTGAFSGYY53 pKa = 9.9FSYY56 pKa = 11.23DD57 pKa = 2.75DD58 pKa = 4.41SYY60 pKa = 11.7FSRR63 pKa = 11.84TDD65 pKa = 3.42GVGGFYY71 pKa = 10.56LVSPDD76 pKa = 3.65SDD78 pKa = 3.62VSLVDD83 pKa = 3.36PTFNAATTNNWDD95 pKa = 3.36YY96 pKa = 11.62DD97 pKa = 3.79LRR99 pKa = 11.84ASDD102 pKa = 3.94SGSLVDD108 pKa = 3.6SLGYY112 pKa = 9.76IAFYY116 pKa = 9.33DD117 pKa = 3.73TYY119 pKa = 10.73DD120 pKa = 3.3TLFRR124 pKa = 11.84TDD126 pKa = 3.17QNDD129 pKa = 3.76DD130 pKa = 3.16VSPNHH135 pKa = 7.01GDD137 pKa = 3.04WTLAAYY143 pKa = 10.38LDD145 pKa = 4.1EE146 pKa = 6.05LEE148 pKa = 5.72NPDD151 pKa = 3.75QNEE154 pKa = 4.3IICIDD159 pKa = 3.44VDD161 pKa = 4.06TLNGASSHH169 pKa = 5.02YY170 pKa = 10.78SKK172 pKa = 10.87LFKK175 pKa = 10.13STLSVEE181 pKa = 3.71RR182 pKa = 11.84WLAGDD187 pKa = 4.1LVYY190 pKa = 10.69ISGLEE195 pKa = 4.08AILEE199 pKa = 4.24NFFTLNDD206 pKa = 3.28KK207 pKa = 10.81RR208 pKa = 11.84LTGNISDD215 pKa = 4.94DD216 pKa = 3.97EE217 pKa = 4.51FTVSCLSVSIAGAPATQEE235 pKa = 4.11LSTLNLLSSLSIPIIQAAPNVGQGNYY261 pKa = 10.19DD262 pKa = 3.07WGSNYY267 pKa = 10.32ADD269 pKa = 4.27VINVGAWNIDD279 pKa = 3.57NEE281 pKa = 4.52DD282 pKa = 3.87EE283 pKa = 4.62LLLSSISTLSTLDD296 pKa = 3.13ILGNGQVLKK305 pKa = 10.65SDD307 pKa = 3.39WGTGSNFGTSFATPKK322 pKa = 10.08VAADD326 pKa = 3.07ITNFYY331 pKa = 10.95SSFISAIEE339 pKa = 4.08SEE341 pKa = 4.48GTSLGEE347 pKa = 4.38AQAQSQALNIDD358 pKa = 3.4YY359 pKa = 10.95SDD361 pKa = 3.7VVNVLLEE368 pKa = 4.17QISTEE373 pKa = 4.44LIVTLDD379 pKa = 3.61LAGNQATYY387 pKa = 9.74HH388 pKa = 5.77PKK390 pKa = 10.11VRR392 pKa = 11.84TSDD395 pKa = 3.67VNVSPTPVKK404 pKa = 9.61ITSTSGGLSGLTVASARR421 pKa = 11.84TNDD424 pKa = 3.27APIFTSGAEE433 pKa = 4.21SVALEE438 pKa = 4.54NITLSSVIYY447 pKa = 10.41NATAEE452 pKa = 4.63DD453 pKa = 3.81SDD455 pKa = 4.52GDD457 pKa = 3.65QLTYY461 pKa = 11.43SLVNSLNILVSGEE474 pKa = 4.24SAVGEE479 pKa = 3.79IDD481 pKa = 3.13IALFDD486 pKa = 5.97DD487 pKa = 4.57ISPLHH492 pKa = 6.59AEE494 pKa = 4.19RR495 pKa = 11.84LGTLAAEE502 pKa = 4.67GAYY505 pKa = 10.45NDD507 pKa = 3.92VVFHH511 pKa = 6.1RR512 pKa = 11.84VIDD515 pKa = 3.87GFMAQTGDD523 pKa = 3.37VQYY526 pKa = 11.26GKK528 pKa = 10.54RR529 pKa = 11.84NNSIEE534 pKa = 3.87NAGRR538 pKa = 11.84GGSNKK543 pKa = 9.02PDD545 pKa = 3.23LVAEE549 pKa = 4.82FSTIPFDD556 pKa = 3.96RR557 pKa = 11.84GIVGMARR564 pKa = 11.84SSDD567 pKa = 3.72PNSANSQFFMMFEE580 pKa = 4.19DD581 pKa = 4.93TYY583 pKa = 11.55SLNGDD588 pKa = 3.66YY589 pKa = 10.77TVVGVVASGLDD600 pKa = 3.37VLDD603 pKa = 4.72LIKK606 pKa = 10.95RR607 pKa = 11.84GDD609 pKa = 3.37ASQNGVVSQNPDD621 pKa = 2.86YY622 pKa = 10.41MEE624 pKa = 4.26EE625 pKa = 4.04VTYY628 pKa = 10.4TPATDD633 pKa = 2.95IFEE636 pKa = 4.7INSTSGEE643 pKa = 3.74ITFINEE649 pKa = 3.86PNFSNMDD656 pKa = 3.56FKK658 pKa = 11.64DD659 pKa = 3.55LVVAVSDD666 pKa = 4.18GSEE669 pKa = 4.01TVYY672 pKa = 9.46KK673 pKa = 10.19TVTLDD678 pKa = 4.7LPPQNITISFSDD690 pKa = 3.4INSVEE695 pKa = 4.13SQVPDD700 pKa = 3.16NDD702 pKa = 4.1LVFKK706 pKa = 10.71GGYY709 pKa = 8.68DD710 pKa = 3.09ATVAVEE716 pKa = 3.85NGKK719 pKa = 10.36LGVLSGEE726 pKa = 3.8ISFTHH731 pKa = 6.57VEE733 pKa = 4.32FASQSYY739 pKa = 10.51DD740 pKa = 3.06HH741 pKa = 7.34GIAISDD747 pKa = 3.73VVLQLRR753 pKa = 11.84DD754 pKa = 3.48IVGLSTLSGNQKK766 pKa = 7.94TAADD770 pKa = 3.7IDD772 pKa = 4.0GDD774 pKa = 4.09GDD776 pKa = 3.91VAISDD781 pKa = 3.82VVSNLRR787 pKa = 11.84HH788 pKa = 5.4IVGLDD793 pKa = 3.48TIEE796 pKa = 3.83QCALVDD802 pKa = 3.61TSDD805 pKa = 3.54QLVTGLTSSTIADD818 pKa = 3.65LTLIQLGDD826 pKa = 3.48VDD828 pKa = 5.48LSATFEE834 pKa = 4.11IAA836 pKa = 3.82
MM1 pKa = 7.77AIKK4 pKa = 10.37SVTVFIDD11 pKa = 3.24QFSDD15 pKa = 3.68RR16 pKa = 11.84IFDD19 pKa = 4.17LGHH22 pKa = 6.82DD23 pKa = 4.02NIYY26 pKa = 10.7LYY28 pKa = 10.49DD29 pKa = 3.67YY30 pKa = 11.33GNFHH34 pKa = 5.93EE35 pKa = 5.29TVWTDD40 pKa = 2.5NYY42 pKa = 10.85YY43 pKa = 10.93NVSTGAFSGYY53 pKa = 9.9FSYY56 pKa = 11.23DD57 pKa = 2.75DD58 pKa = 4.41SYY60 pKa = 11.7FSRR63 pKa = 11.84TDD65 pKa = 3.42GVGGFYY71 pKa = 10.56LVSPDD76 pKa = 3.65SDD78 pKa = 3.62VSLVDD83 pKa = 3.36PTFNAATTNNWDD95 pKa = 3.36YY96 pKa = 11.62DD97 pKa = 3.79LRR99 pKa = 11.84ASDD102 pKa = 3.94SGSLVDD108 pKa = 3.6SLGYY112 pKa = 9.76IAFYY116 pKa = 9.33DD117 pKa = 3.73TYY119 pKa = 10.73DD120 pKa = 3.3TLFRR124 pKa = 11.84TDD126 pKa = 3.17QNDD129 pKa = 3.76DD130 pKa = 3.16VSPNHH135 pKa = 7.01GDD137 pKa = 3.04WTLAAYY143 pKa = 10.38LDD145 pKa = 4.1EE146 pKa = 6.05LEE148 pKa = 5.72NPDD151 pKa = 3.75QNEE154 pKa = 4.3IICIDD159 pKa = 3.44VDD161 pKa = 4.06TLNGASSHH169 pKa = 5.02YY170 pKa = 10.78SKK172 pKa = 10.87LFKK175 pKa = 10.13STLSVEE181 pKa = 3.71RR182 pKa = 11.84WLAGDD187 pKa = 4.1LVYY190 pKa = 10.69ISGLEE195 pKa = 4.08AILEE199 pKa = 4.24NFFTLNDD206 pKa = 3.28KK207 pKa = 10.81RR208 pKa = 11.84LTGNISDD215 pKa = 4.94DD216 pKa = 3.97EE217 pKa = 4.51FTVSCLSVSIAGAPATQEE235 pKa = 4.11LSTLNLLSSLSIPIIQAAPNVGQGNYY261 pKa = 10.19DD262 pKa = 3.07WGSNYY267 pKa = 10.32ADD269 pKa = 4.27VINVGAWNIDD279 pKa = 3.57NEE281 pKa = 4.52DD282 pKa = 3.87EE283 pKa = 4.62LLLSSISTLSTLDD296 pKa = 3.13ILGNGQVLKK305 pKa = 10.65SDD307 pKa = 3.39WGTGSNFGTSFATPKK322 pKa = 10.08VAADD326 pKa = 3.07ITNFYY331 pKa = 10.95SSFISAIEE339 pKa = 4.08SEE341 pKa = 4.48GTSLGEE347 pKa = 4.38AQAQSQALNIDD358 pKa = 3.4YY359 pKa = 10.95SDD361 pKa = 3.7VVNVLLEE368 pKa = 4.17QISTEE373 pKa = 4.44LIVTLDD379 pKa = 3.61LAGNQATYY387 pKa = 9.74HH388 pKa = 5.77PKK390 pKa = 10.11VRR392 pKa = 11.84TSDD395 pKa = 3.67VNVSPTPVKK404 pKa = 9.61ITSTSGGLSGLTVASARR421 pKa = 11.84TNDD424 pKa = 3.27APIFTSGAEE433 pKa = 4.21SVALEE438 pKa = 4.54NITLSSVIYY447 pKa = 10.41NATAEE452 pKa = 4.63DD453 pKa = 3.81SDD455 pKa = 4.52GDD457 pKa = 3.65QLTYY461 pKa = 11.43SLVNSLNILVSGEE474 pKa = 4.24SAVGEE479 pKa = 3.79IDD481 pKa = 3.13IALFDD486 pKa = 5.97DD487 pKa = 4.57ISPLHH492 pKa = 6.59AEE494 pKa = 4.19RR495 pKa = 11.84LGTLAAEE502 pKa = 4.67GAYY505 pKa = 10.45NDD507 pKa = 3.92VVFHH511 pKa = 6.1RR512 pKa = 11.84VIDD515 pKa = 3.87GFMAQTGDD523 pKa = 3.37VQYY526 pKa = 11.26GKK528 pKa = 10.54RR529 pKa = 11.84NNSIEE534 pKa = 3.87NAGRR538 pKa = 11.84GGSNKK543 pKa = 9.02PDD545 pKa = 3.23LVAEE549 pKa = 4.82FSTIPFDD556 pKa = 3.96RR557 pKa = 11.84GIVGMARR564 pKa = 11.84SSDD567 pKa = 3.72PNSANSQFFMMFEE580 pKa = 4.19DD581 pKa = 4.93TYY583 pKa = 11.55SLNGDD588 pKa = 3.66YY589 pKa = 10.77TVVGVVASGLDD600 pKa = 3.37VLDD603 pKa = 4.72LIKK606 pKa = 10.95RR607 pKa = 11.84GDD609 pKa = 3.37ASQNGVVSQNPDD621 pKa = 2.86YY622 pKa = 10.41MEE624 pKa = 4.26EE625 pKa = 4.04VTYY628 pKa = 10.4TPATDD633 pKa = 2.95IFEE636 pKa = 4.7INSTSGEE643 pKa = 3.74ITFINEE649 pKa = 3.86PNFSNMDD656 pKa = 3.56FKK658 pKa = 11.64DD659 pKa = 3.55LVVAVSDD666 pKa = 4.18GSEE669 pKa = 4.01TVYY672 pKa = 9.46KK673 pKa = 10.19TVTLDD678 pKa = 4.7LPPQNITISFSDD690 pKa = 3.4INSVEE695 pKa = 4.13SQVPDD700 pKa = 3.16NDD702 pKa = 4.1LVFKK706 pKa = 10.71GGYY709 pKa = 8.68DD710 pKa = 3.09ATVAVEE716 pKa = 3.85NGKK719 pKa = 10.36LGVLSGEE726 pKa = 3.8ISFTHH731 pKa = 6.57VEE733 pKa = 4.32FASQSYY739 pKa = 10.51DD740 pKa = 3.06HH741 pKa = 7.34GIAISDD747 pKa = 3.73VVLQLRR753 pKa = 11.84DD754 pKa = 3.48IVGLSTLSGNQKK766 pKa = 7.94TAADD770 pKa = 3.7IDD772 pKa = 4.0GDD774 pKa = 4.09GDD776 pKa = 3.91VAISDD781 pKa = 3.82VVSNLRR787 pKa = 11.84HH788 pKa = 5.4IVGLDD793 pKa = 3.48TIEE796 pKa = 3.83QCALVDD802 pKa = 3.61TSDD805 pKa = 3.54QLVTGLTSSTIADD818 pKa = 3.65LTLIQLGDD826 pKa = 3.48VDD828 pKa = 5.48LSATFEE834 pKa = 4.11IAA836 pKa = 3.82
Molecular weight: 89.83 kDa
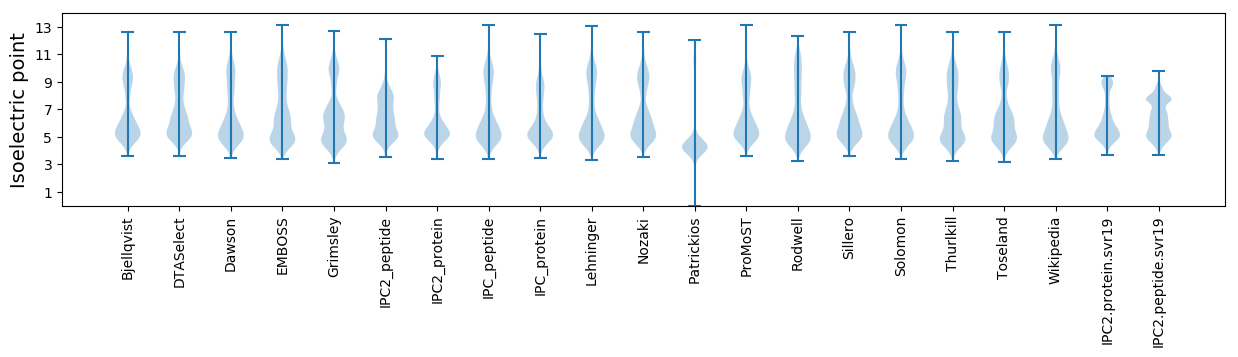
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A177HCU2|A0A177HCU2_9RHOB Phytoene desaturase (Neurosporene-forming) OS=Rhodobacteraceae bacterium SB2 OX=1689867 GN=crtI PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.43EE41 pKa = 3.72LSAA44 pKa = 5.03
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.43EE41 pKa = 3.72LSAA44 pKa = 5.03
Molecular weight: 5.18 kDa
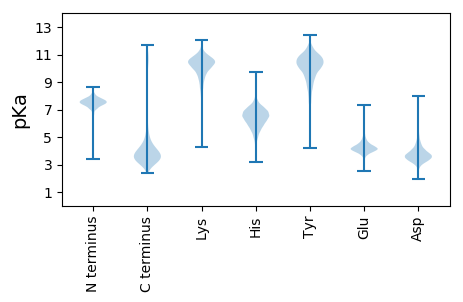
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1066040 |
29 |
4255 |
305.5 |
33.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.136 ± 0.054 | 1.049 ± 0.014 |
5.608 ± 0.047 | 5.65 ± 0.036 |
4.17 ± 0.03 | 8.006 ± 0.05 |
2.12 ± 0.022 | 6.032 ± 0.033 |
4.177 ± 0.031 | 10.31 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.765 ± 0.022 | 3.313 ± 0.029 |
4.578 ± 0.029 | 3.921 ± 0.026 |
5.494 ± 0.036 | 6.104 ± 0.042 |
5.159 ± 0.037 | 6.61 ± 0.033 |
1.338 ± 0.017 | 2.46 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
