
[Candida] intermedia
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Saccharomycotina; Saccharomycetes; Saccharomycetales; Metschnikowiaceae; Clavispora; Clavispora/Candida clade
Average proteome isoelectric point is 6.24
Get precalculated fractions of proteins
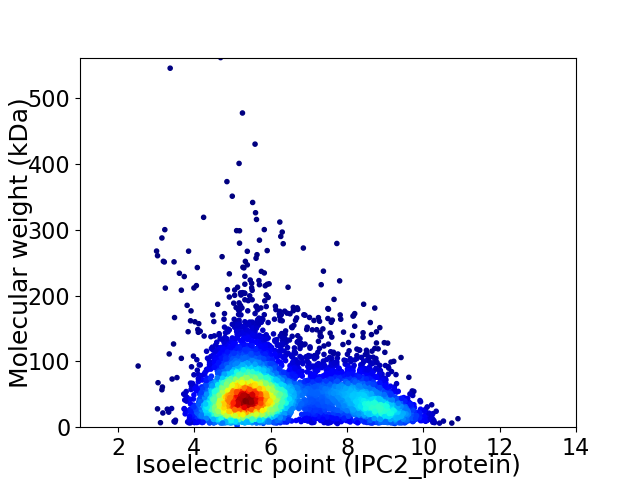
Virtual 2D-PAGE plot for 5921 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L0CSD2|A0A1L0CSD2_9ASCO DNA polymerase OS=[Candida] intermedia OX=45354 GN=SAMEA4029010_CIC11G00000002857 PE=3 SV=1
MM1 pKa = 7.59ILHH4 pKa = 6.57EE5 pKa = 4.55FVSLALLATVVQAATTTVSTTSTFPFTSYY34 pKa = 10.52SQSSTDD40 pKa = 2.79QWLIEE45 pKa = 4.25TDD47 pKa = 3.51GSLTFSSILNLKK59 pKa = 7.91TGKK62 pKa = 9.8NFEE65 pKa = 4.41NYY67 pKa = 10.62GDD69 pKa = 4.04VIMNANPGATLEE81 pKa = 4.19MDD83 pKa = 3.46SLTNYY88 pKa = 7.24EE89 pKa = 4.16TGSVKK94 pKa = 10.11IAASGGSLTFQSSNTYY110 pKa = 8.83TNQGLWVMSTPGDD123 pKa = 3.9LEE125 pKa = 4.11VDD127 pKa = 3.92LTDD130 pKa = 4.85SLYY133 pKa = 10.68TLLNSGSLTYY143 pKa = 10.64EE144 pKa = 3.85SGGDD148 pKa = 3.42MAISVSKK155 pKa = 10.68VFINSEE161 pKa = 4.32EE162 pKa = 4.16VSFTASGKK170 pKa = 8.58GTFSFSSDD178 pKa = 3.36FTNSGTMIVTSSSVSDD194 pKa = 3.37TFKK197 pKa = 10.94ISDD200 pKa = 3.53TFVNDD205 pKa = 2.78NYY207 pKa = 10.67FVYY210 pKa = 10.54NYY212 pKa = 10.18GLNSGTAFSLSSLLSTYY229 pKa = 9.6TNNGIMNIYY238 pKa = 10.23GSTGSTNMNQDD249 pKa = 3.1GPIVNDD255 pKa = 3.77GSICLRR261 pKa = 11.84HH262 pKa = 4.4TTYY265 pKa = 11.24NGGAKK270 pKa = 10.43LSGTGCWVLADD281 pKa = 3.69SSVFTQHH288 pKa = 5.84VARR291 pKa = 11.84LANTQTIVFNDD302 pKa = 3.85TSASIRR308 pKa = 11.84VSSVGLGSHH317 pKa = 4.27YY318 pKa = 9.85TIYY321 pKa = 10.71GVQNGMTFLTSSLLIGSVVYY341 pKa = 10.86SPFTGILTLTLGLLSTTYY359 pKa = 10.61DD360 pKa = 2.67IGLGYY365 pKa = 10.51DD366 pKa = 3.29PTGFTVSSNNVRR378 pKa = 11.84YY379 pKa = 10.2VGNDD383 pKa = 3.12PVKK386 pKa = 10.43RR387 pKa = 11.84DD388 pKa = 3.36IPGEE392 pKa = 4.28CICEE396 pKa = 4.03PQEE399 pKa = 3.86QSSSVFSSSASSSSSISEE417 pKa = 4.21TSSEE421 pKa = 4.14ATSSQSSEE429 pKa = 3.89ATSSQSSEE437 pKa = 3.91VSSSQSSEE445 pKa = 3.75ASSSQSSEE453 pKa = 3.72ASSSQSSEE461 pKa = 3.72ASSSQSSEE469 pKa = 3.72ASSSQSSEE477 pKa = 3.78ATSSQSSEE485 pKa = 3.82ASSSQSSEE493 pKa = 3.72ASSSQSSEE501 pKa = 3.72ASSSQSSEE509 pKa = 3.72ASSSQSSEE517 pKa = 3.72ASSSQSSEE525 pKa = 3.72ASSSQSSEE533 pKa = 3.8AASSQSSEE541 pKa = 4.03ALSSQSSEE549 pKa = 4.07ASPQSSEE556 pKa = 4.07ASSSHH561 pKa = 6.62PASASNSQTEE571 pKa = 4.05QSTAPSVQTSQSVEE585 pKa = 3.61EE586 pKa = 4.29SQALNSNTEE595 pKa = 4.07YY596 pKa = 7.78TTTWTTTNSDD606 pKa = 3.26GSVEE610 pKa = 4.08TDD612 pKa = 2.95SGVVIVTTDD621 pKa = 3.01SAGVLTTSTSKK632 pKa = 10.87LSPAQYY638 pKa = 6.53TTTWTTTNSDD648 pKa = 3.26GSVEE652 pKa = 4.08TDD654 pKa = 2.96SGVVFVTTDD663 pKa = 2.95SAGVLTTSTSKK674 pKa = 10.87LSPAQYY680 pKa = 6.53TTTWTTTNSDD690 pKa = 3.26GSVEE694 pKa = 4.08TDD696 pKa = 2.95SGVVIVTTDD705 pKa = 3.01SAGVLTTSTSKK716 pKa = 10.87LSPAQYY722 pKa = 6.53TTTWTTTNSDD732 pKa = 3.26GSVEE736 pKa = 4.08TDD738 pKa = 2.95SGVVIVTTDD747 pKa = 3.01SAGVLTTSTSKK758 pKa = 10.66LAPAQYY764 pKa = 6.91TTTWTTTNSDD774 pKa = 3.26GSVEE778 pKa = 4.08TDD780 pKa = 2.96SGVVFVTTDD789 pKa = 2.95SAGVLTTSTSKK800 pKa = 10.87LSPAQYY806 pKa = 6.53TTTWTTTNSDD816 pKa = 3.26GSVEE820 pKa = 4.08TDD822 pKa = 2.95SGVVIVTTDD831 pKa = 3.01SAGVLTTSTSKK842 pKa = 10.9LSPAKK847 pKa = 9.75YY848 pKa = 5.3TTTWTTTNSDD858 pKa = 3.26GSVEE862 pKa = 4.08TDD864 pKa = 2.96SGVVFVTTDD873 pKa = 2.95SAGVLTTSTSKK884 pKa = 10.87LSPAQYY890 pKa = 6.53TTTWTTTNSDD900 pKa = 3.26GSVEE904 pKa = 4.08TDD906 pKa = 2.95SGVVIVTTDD915 pKa = 3.01SAGVLTTSTSKK926 pKa = 10.66LAPAQYY932 pKa = 6.91TTTWTTTNSDD942 pKa = 3.26GSVEE946 pKa = 4.08TDD948 pKa = 2.95SGVVIVTTDD957 pKa = 3.01SAGVLTTSTSKK968 pKa = 10.66LAPAQYY974 pKa = 6.91TTTWTTTNSDD984 pKa = 3.26GSVEE988 pKa = 4.08TDD990 pKa = 2.96SGVVFVTTDD999 pKa = 2.95SAGVLTTSTSKK1010 pKa = 10.66LAPAQYY1016 pKa = 6.91TTTWTTTNSDD1026 pKa = 3.26GSVEE1030 pKa = 4.08TDD1032 pKa = 2.95SGVVIVTTDD1041 pKa = 3.01SAGVLTTSTSKK1052 pKa = 10.66LAPAQYY1058 pKa = 6.91TTTWTTTNSDD1068 pKa = 3.26GSVEE1072 pKa = 4.08TDD1074 pKa = 2.96SGVVFVTTDD1083 pKa = 2.95SAGVLTTSTSKK1094 pKa = 10.66LAPAQYY1100 pKa = 6.91TTTWTTTNSDD1110 pKa = 3.26GSVEE1114 pKa = 4.08TDD1116 pKa = 2.95SGVVIVTTDD1125 pKa = 3.01SAGVLTTSTSKK1136 pKa = 10.87LSPAQYY1142 pKa = 6.53TTTWTTTNSDD1152 pKa = 3.26GSVEE1156 pKa = 4.08TDD1158 pKa = 2.96SGVVFVTTDD1167 pKa = 2.95SAGVLTTSTSKK1178 pKa = 10.66LAPAQYY1184 pKa = 6.91TTTWTTTNSDD1194 pKa = 3.26GSVEE1198 pKa = 4.08TDD1200 pKa = 2.96SGVVFVTTDD1209 pKa = 2.95SAGVLTTSTSKK1220 pKa = 10.9LSPAKK1225 pKa = 9.75YY1226 pKa = 5.3TTTWTTTNSDD1236 pKa = 3.26GSVEE1240 pKa = 4.08TDD1242 pKa = 2.96SGVVFVTTDD1251 pKa = 2.95SAGVLTTSTSKK1262 pKa = 10.87LSPAQYY1268 pKa = 6.53TTTWTTTNSDD1278 pKa = 3.26GSVEE1282 pKa = 4.08TDD1284 pKa = 2.95SGVVIVTTDD1293 pKa = 3.01SAGVLTTSTSKK1304 pKa = 10.66LAPAQYY1310 pKa = 6.91TTTWTTTNSDD1320 pKa = 3.26GSVEE1324 pKa = 4.08TDD1326 pKa = 2.95SGVVIVTTDD1335 pKa = 2.98SAGVFTTSTSKK1346 pKa = 10.66LAPAQYY1352 pKa = 6.91TTTWTTTNSDD1362 pKa = 3.26GSVEE1366 pKa = 4.08TDD1368 pKa = 2.96SGVVFVTTDD1377 pKa = 2.95SAGVLTTSTSKK1388 pKa = 10.66LAPAQYY1394 pKa = 6.91TTTWTTTNSDD1404 pKa = 3.26GSVEE1408 pKa = 4.08TDD1410 pKa = 2.95SGVVIVTTDD1419 pKa = 3.01SAGVLTTSTSKK1430 pKa = 10.87LSPAQYY1436 pKa = 6.53TTTWTTTNSDD1446 pKa = 3.26GSVEE1450 pKa = 4.08TDD1452 pKa = 2.95SGVVIVTTDD1461 pKa = 3.01SAGVLTTSTSKK1472 pKa = 10.66LAPAQYY1478 pKa = 6.91TTTWTTTNSDD1488 pKa = 3.26GSVEE1492 pKa = 4.08TDD1494 pKa = 2.95SGVVIVTTDD1503 pKa = 3.01SAGVLTTSTSKK1514 pKa = 10.66LAPAQYY1520 pKa = 6.91TTTWTTTNSDD1530 pKa = 3.26GSVEE1534 pKa = 4.08TDD1536 pKa = 2.96SGVVFVTTDD1545 pKa = 2.95SAGVLTTSTSKK1556 pKa = 10.66LAPAQYY1562 pKa = 6.91TTTWTTTNSDD1572 pKa = 3.26GSVEE1576 pKa = 4.08TDD1578 pKa = 2.95SGVVIVTTDD1587 pKa = 3.01SAGVLTTSTSKK1598 pKa = 10.87LSPAQYY1604 pKa = 6.53TTTWTTTNSDD1614 pKa = 3.26GSVEE1618 pKa = 4.08TDD1620 pKa = 2.96SGVVFVTTDD1629 pKa = 2.95SAGVLTTSTSKK1640 pKa = 10.66LAPAQYY1646 pKa = 6.91TTTWTTTNSDD1656 pKa = 3.26GSVEE1660 pKa = 4.08TDD1662 pKa = 2.95SGVVIVTTDD1671 pKa = 3.01SAGVLTTSTSKK1682 pKa = 10.66LAPAQYY1688 pKa = 6.91TTTWTTTNSDD1698 pKa = 3.26GSVEE1702 pKa = 4.08TDD1704 pKa = 2.96SGVVFVTTDD1713 pKa = 2.95SAGVLTTSTSKK1724 pKa = 10.66LAPAQYY1730 pKa = 6.91TTTWTTTNSDD1740 pKa = 3.26GSVEE1744 pKa = 4.08TDD1746 pKa = 2.96SGVVFVTTDD1755 pKa = 2.95SAGVLTTSTSKK1766 pKa = 10.57LAPASEE1772 pKa = 4.59VTSSGSSTISAEE1784 pKa = 4.05PTSLSTASPLINPAGLVGLDD1804 pKa = 3.86IITDD1808 pKa = 3.75ISVHH1812 pKa = 6.44DD1813 pKa = 4.16NSTGSFIGYY1822 pKa = 9.64KK1823 pKa = 10.06DD1824 pKa = 3.81DD1825 pKa = 4.23LFTVVVTLKK1834 pKa = 10.31SASSIPLGSITQFTVPEE1851 pKa = 4.15VFYY1854 pKa = 11.45GNLNPFEE1861 pKa = 4.59VYY1863 pKa = 10.55SGDD1866 pKa = 3.49GEE1868 pKa = 4.67YY1869 pKa = 10.93VGTISSDD1876 pKa = 3.26DD1877 pKa = 3.95SNVFTIVFDD1886 pKa = 4.19GKK1888 pKa = 10.84FGLTHH1893 pKa = 6.5TDD1895 pKa = 2.47IEE1897 pKa = 4.32AAFTFNVKK1905 pKa = 9.73LRR1907 pKa = 11.84YY1908 pKa = 6.26TTALTKK1914 pKa = 10.32RR1915 pKa = 11.84AEE1917 pKa = 4.07LSLDD1921 pKa = 3.54LSRR1924 pKa = 11.84PVAQPFVFTTVSSAFTSIIFFPDD1947 pKa = 2.98VSLDD1951 pKa = 4.0GIVGKK1956 pKa = 9.61VLIGDD1961 pKa = 3.81VYY1963 pKa = 11.43VFVGDD1968 pKa = 4.16ALSGASNSAISTGYY1982 pKa = 9.84SAPFSYY1988 pKa = 10.25WNNTVTATEE1997 pKa = 4.39TGTSLVTVTSCANGNCHH2014 pKa = 5.87GTSKK2018 pKa = 9.61LTSKK2022 pKa = 10.65ASSAVAPTTVSVTKK2036 pKa = 10.23DD2037 pKa = 2.93LTVIVTITSCSNHH2050 pKa = 4.77VCVPTTSKK2058 pKa = 11.19ALASIATTTVHH2069 pKa = 6.52GEE2071 pKa = 3.97VVLYY2075 pKa = 7.42TTYY2078 pKa = 10.89CPLSSEE2084 pKa = 4.37TGEE2087 pKa = 4.18KK2088 pKa = 10.05AAQEE2092 pKa = 4.4TSGSANSGSNSGSNAGSANAGSNAGSANAGSNSGSNAGSNSGSNAGSNSGSNSGSNAGSNSGSNAGSNSGSNAGSNSGSNSGSNAGSANAGSNVGSNAASGANVQYY2198 pKa = 9.43DD2199 pKa = 3.74TGISTLITATKK2210 pKa = 8.9STSSTSGNTGPAVSVNEE2227 pKa = 3.7NSARR2231 pKa = 11.84GSVNTSSFTAIFIGLLSLLTTFVV2254 pKa = 3.24
MM1 pKa = 7.59ILHH4 pKa = 6.57EE5 pKa = 4.55FVSLALLATVVQAATTTVSTTSTFPFTSYY34 pKa = 10.52SQSSTDD40 pKa = 2.79QWLIEE45 pKa = 4.25TDD47 pKa = 3.51GSLTFSSILNLKK59 pKa = 7.91TGKK62 pKa = 9.8NFEE65 pKa = 4.41NYY67 pKa = 10.62GDD69 pKa = 4.04VIMNANPGATLEE81 pKa = 4.19MDD83 pKa = 3.46SLTNYY88 pKa = 7.24EE89 pKa = 4.16TGSVKK94 pKa = 10.11IAASGGSLTFQSSNTYY110 pKa = 8.83TNQGLWVMSTPGDD123 pKa = 3.9LEE125 pKa = 4.11VDD127 pKa = 3.92LTDD130 pKa = 4.85SLYY133 pKa = 10.68TLLNSGSLTYY143 pKa = 10.64EE144 pKa = 3.85SGGDD148 pKa = 3.42MAISVSKK155 pKa = 10.68VFINSEE161 pKa = 4.32EE162 pKa = 4.16VSFTASGKK170 pKa = 8.58GTFSFSSDD178 pKa = 3.36FTNSGTMIVTSSSVSDD194 pKa = 3.37TFKK197 pKa = 10.94ISDD200 pKa = 3.53TFVNDD205 pKa = 2.78NYY207 pKa = 10.67FVYY210 pKa = 10.54NYY212 pKa = 10.18GLNSGTAFSLSSLLSTYY229 pKa = 9.6TNNGIMNIYY238 pKa = 10.23GSTGSTNMNQDD249 pKa = 3.1GPIVNDD255 pKa = 3.77GSICLRR261 pKa = 11.84HH262 pKa = 4.4TTYY265 pKa = 11.24NGGAKK270 pKa = 10.43LSGTGCWVLADD281 pKa = 3.69SSVFTQHH288 pKa = 5.84VARR291 pKa = 11.84LANTQTIVFNDD302 pKa = 3.85TSASIRR308 pKa = 11.84VSSVGLGSHH317 pKa = 4.27YY318 pKa = 9.85TIYY321 pKa = 10.71GVQNGMTFLTSSLLIGSVVYY341 pKa = 10.86SPFTGILTLTLGLLSTTYY359 pKa = 10.61DD360 pKa = 2.67IGLGYY365 pKa = 10.51DD366 pKa = 3.29PTGFTVSSNNVRR378 pKa = 11.84YY379 pKa = 10.2VGNDD383 pKa = 3.12PVKK386 pKa = 10.43RR387 pKa = 11.84DD388 pKa = 3.36IPGEE392 pKa = 4.28CICEE396 pKa = 4.03PQEE399 pKa = 3.86QSSSVFSSSASSSSSISEE417 pKa = 4.21TSSEE421 pKa = 4.14ATSSQSSEE429 pKa = 3.89ATSSQSSEE437 pKa = 3.91VSSSQSSEE445 pKa = 3.75ASSSQSSEE453 pKa = 3.72ASSSQSSEE461 pKa = 3.72ASSSQSSEE469 pKa = 3.72ASSSQSSEE477 pKa = 3.78ATSSQSSEE485 pKa = 3.82ASSSQSSEE493 pKa = 3.72ASSSQSSEE501 pKa = 3.72ASSSQSSEE509 pKa = 3.72ASSSQSSEE517 pKa = 3.72ASSSQSSEE525 pKa = 3.72ASSSQSSEE533 pKa = 3.8AASSQSSEE541 pKa = 4.03ALSSQSSEE549 pKa = 4.07ASPQSSEE556 pKa = 4.07ASSSHH561 pKa = 6.62PASASNSQTEE571 pKa = 4.05QSTAPSVQTSQSVEE585 pKa = 3.61EE586 pKa = 4.29SQALNSNTEE595 pKa = 4.07YY596 pKa = 7.78TTTWTTTNSDD606 pKa = 3.26GSVEE610 pKa = 4.08TDD612 pKa = 2.95SGVVIVTTDD621 pKa = 3.01SAGVLTTSTSKK632 pKa = 10.87LSPAQYY638 pKa = 6.53TTTWTTTNSDD648 pKa = 3.26GSVEE652 pKa = 4.08TDD654 pKa = 2.96SGVVFVTTDD663 pKa = 2.95SAGVLTTSTSKK674 pKa = 10.87LSPAQYY680 pKa = 6.53TTTWTTTNSDD690 pKa = 3.26GSVEE694 pKa = 4.08TDD696 pKa = 2.95SGVVIVTTDD705 pKa = 3.01SAGVLTTSTSKK716 pKa = 10.87LSPAQYY722 pKa = 6.53TTTWTTTNSDD732 pKa = 3.26GSVEE736 pKa = 4.08TDD738 pKa = 2.95SGVVIVTTDD747 pKa = 3.01SAGVLTTSTSKK758 pKa = 10.66LAPAQYY764 pKa = 6.91TTTWTTTNSDD774 pKa = 3.26GSVEE778 pKa = 4.08TDD780 pKa = 2.96SGVVFVTTDD789 pKa = 2.95SAGVLTTSTSKK800 pKa = 10.87LSPAQYY806 pKa = 6.53TTTWTTTNSDD816 pKa = 3.26GSVEE820 pKa = 4.08TDD822 pKa = 2.95SGVVIVTTDD831 pKa = 3.01SAGVLTTSTSKK842 pKa = 10.9LSPAKK847 pKa = 9.75YY848 pKa = 5.3TTTWTTTNSDD858 pKa = 3.26GSVEE862 pKa = 4.08TDD864 pKa = 2.96SGVVFVTTDD873 pKa = 2.95SAGVLTTSTSKK884 pKa = 10.87LSPAQYY890 pKa = 6.53TTTWTTTNSDD900 pKa = 3.26GSVEE904 pKa = 4.08TDD906 pKa = 2.95SGVVIVTTDD915 pKa = 3.01SAGVLTTSTSKK926 pKa = 10.66LAPAQYY932 pKa = 6.91TTTWTTTNSDD942 pKa = 3.26GSVEE946 pKa = 4.08TDD948 pKa = 2.95SGVVIVTTDD957 pKa = 3.01SAGVLTTSTSKK968 pKa = 10.66LAPAQYY974 pKa = 6.91TTTWTTTNSDD984 pKa = 3.26GSVEE988 pKa = 4.08TDD990 pKa = 2.96SGVVFVTTDD999 pKa = 2.95SAGVLTTSTSKK1010 pKa = 10.66LAPAQYY1016 pKa = 6.91TTTWTTTNSDD1026 pKa = 3.26GSVEE1030 pKa = 4.08TDD1032 pKa = 2.95SGVVIVTTDD1041 pKa = 3.01SAGVLTTSTSKK1052 pKa = 10.66LAPAQYY1058 pKa = 6.91TTTWTTTNSDD1068 pKa = 3.26GSVEE1072 pKa = 4.08TDD1074 pKa = 2.96SGVVFVTTDD1083 pKa = 2.95SAGVLTTSTSKK1094 pKa = 10.66LAPAQYY1100 pKa = 6.91TTTWTTTNSDD1110 pKa = 3.26GSVEE1114 pKa = 4.08TDD1116 pKa = 2.95SGVVIVTTDD1125 pKa = 3.01SAGVLTTSTSKK1136 pKa = 10.87LSPAQYY1142 pKa = 6.53TTTWTTTNSDD1152 pKa = 3.26GSVEE1156 pKa = 4.08TDD1158 pKa = 2.96SGVVFVTTDD1167 pKa = 2.95SAGVLTTSTSKK1178 pKa = 10.66LAPAQYY1184 pKa = 6.91TTTWTTTNSDD1194 pKa = 3.26GSVEE1198 pKa = 4.08TDD1200 pKa = 2.96SGVVFVTTDD1209 pKa = 2.95SAGVLTTSTSKK1220 pKa = 10.9LSPAKK1225 pKa = 9.75YY1226 pKa = 5.3TTTWTTTNSDD1236 pKa = 3.26GSVEE1240 pKa = 4.08TDD1242 pKa = 2.96SGVVFVTTDD1251 pKa = 2.95SAGVLTTSTSKK1262 pKa = 10.87LSPAQYY1268 pKa = 6.53TTTWTTTNSDD1278 pKa = 3.26GSVEE1282 pKa = 4.08TDD1284 pKa = 2.95SGVVIVTTDD1293 pKa = 3.01SAGVLTTSTSKK1304 pKa = 10.66LAPAQYY1310 pKa = 6.91TTTWTTTNSDD1320 pKa = 3.26GSVEE1324 pKa = 4.08TDD1326 pKa = 2.95SGVVIVTTDD1335 pKa = 2.98SAGVFTTSTSKK1346 pKa = 10.66LAPAQYY1352 pKa = 6.91TTTWTTTNSDD1362 pKa = 3.26GSVEE1366 pKa = 4.08TDD1368 pKa = 2.96SGVVFVTTDD1377 pKa = 2.95SAGVLTTSTSKK1388 pKa = 10.66LAPAQYY1394 pKa = 6.91TTTWTTTNSDD1404 pKa = 3.26GSVEE1408 pKa = 4.08TDD1410 pKa = 2.95SGVVIVTTDD1419 pKa = 3.01SAGVLTTSTSKK1430 pKa = 10.87LSPAQYY1436 pKa = 6.53TTTWTTTNSDD1446 pKa = 3.26GSVEE1450 pKa = 4.08TDD1452 pKa = 2.95SGVVIVTTDD1461 pKa = 3.01SAGVLTTSTSKK1472 pKa = 10.66LAPAQYY1478 pKa = 6.91TTTWTTTNSDD1488 pKa = 3.26GSVEE1492 pKa = 4.08TDD1494 pKa = 2.95SGVVIVTTDD1503 pKa = 3.01SAGVLTTSTSKK1514 pKa = 10.66LAPAQYY1520 pKa = 6.91TTTWTTTNSDD1530 pKa = 3.26GSVEE1534 pKa = 4.08TDD1536 pKa = 2.96SGVVFVTTDD1545 pKa = 2.95SAGVLTTSTSKK1556 pKa = 10.66LAPAQYY1562 pKa = 6.91TTTWTTTNSDD1572 pKa = 3.26GSVEE1576 pKa = 4.08TDD1578 pKa = 2.95SGVVIVTTDD1587 pKa = 3.01SAGVLTTSTSKK1598 pKa = 10.87LSPAQYY1604 pKa = 6.53TTTWTTTNSDD1614 pKa = 3.26GSVEE1618 pKa = 4.08TDD1620 pKa = 2.96SGVVFVTTDD1629 pKa = 2.95SAGVLTTSTSKK1640 pKa = 10.66LAPAQYY1646 pKa = 6.91TTTWTTTNSDD1656 pKa = 3.26GSVEE1660 pKa = 4.08TDD1662 pKa = 2.95SGVVIVTTDD1671 pKa = 3.01SAGVLTTSTSKK1682 pKa = 10.66LAPAQYY1688 pKa = 6.91TTTWTTTNSDD1698 pKa = 3.26GSVEE1702 pKa = 4.08TDD1704 pKa = 2.96SGVVFVTTDD1713 pKa = 2.95SAGVLTTSTSKK1724 pKa = 10.66LAPAQYY1730 pKa = 6.91TTTWTTTNSDD1740 pKa = 3.26GSVEE1744 pKa = 4.08TDD1746 pKa = 2.96SGVVFVTTDD1755 pKa = 2.95SAGVLTTSTSKK1766 pKa = 10.57LAPASEE1772 pKa = 4.59VTSSGSSTISAEE1784 pKa = 4.05PTSLSTASPLINPAGLVGLDD1804 pKa = 3.86IITDD1808 pKa = 3.75ISVHH1812 pKa = 6.44DD1813 pKa = 4.16NSTGSFIGYY1822 pKa = 9.64KK1823 pKa = 10.06DD1824 pKa = 3.81DD1825 pKa = 4.23LFTVVVTLKK1834 pKa = 10.31SASSIPLGSITQFTVPEE1851 pKa = 4.15VFYY1854 pKa = 11.45GNLNPFEE1861 pKa = 4.59VYY1863 pKa = 10.55SGDD1866 pKa = 3.49GEE1868 pKa = 4.67YY1869 pKa = 10.93VGTISSDD1876 pKa = 3.26DD1877 pKa = 3.95SNVFTIVFDD1886 pKa = 4.19GKK1888 pKa = 10.84FGLTHH1893 pKa = 6.5TDD1895 pKa = 2.47IEE1897 pKa = 4.32AAFTFNVKK1905 pKa = 9.73LRR1907 pKa = 11.84YY1908 pKa = 6.26TTALTKK1914 pKa = 10.32RR1915 pKa = 11.84AEE1917 pKa = 4.07LSLDD1921 pKa = 3.54LSRR1924 pKa = 11.84PVAQPFVFTTVSSAFTSIIFFPDD1947 pKa = 2.98VSLDD1951 pKa = 4.0GIVGKK1956 pKa = 9.61VLIGDD1961 pKa = 3.81VYY1963 pKa = 11.43VFVGDD1968 pKa = 4.16ALSGASNSAISTGYY1982 pKa = 9.84SAPFSYY1988 pKa = 10.25WNNTVTATEE1997 pKa = 4.39TGTSLVTVTSCANGNCHH2014 pKa = 5.87GTSKK2018 pKa = 9.61LTSKK2022 pKa = 10.65ASSAVAPTTVSVTKK2036 pKa = 10.23DD2037 pKa = 2.93LTVIVTITSCSNHH2050 pKa = 4.77VCVPTTSKK2058 pKa = 11.19ALASIATTTVHH2069 pKa = 6.52GEE2071 pKa = 3.97VVLYY2075 pKa = 7.42TTYY2078 pKa = 10.89CPLSSEE2084 pKa = 4.37TGEE2087 pKa = 4.18KK2088 pKa = 10.05AAQEE2092 pKa = 4.4TSGSANSGSNSGSNAGSANAGSNAGSANAGSNSGSNAGSNSGSNAGSNSGSNSGSNAGSNSGSNAGSNSGSNAGSNSGSNSGSNAGSANAGSNVGSNAASGANVQYY2198 pKa = 9.43DD2199 pKa = 3.74TGISTLITATKK2210 pKa = 8.9STSSTSGNTGPAVSVNEE2227 pKa = 3.7NSARR2231 pKa = 11.84GSVNTSSFTAIFIGLLSLLTTFVV2254 pKa = 3.24
Molecular weight: 229.06 kDa
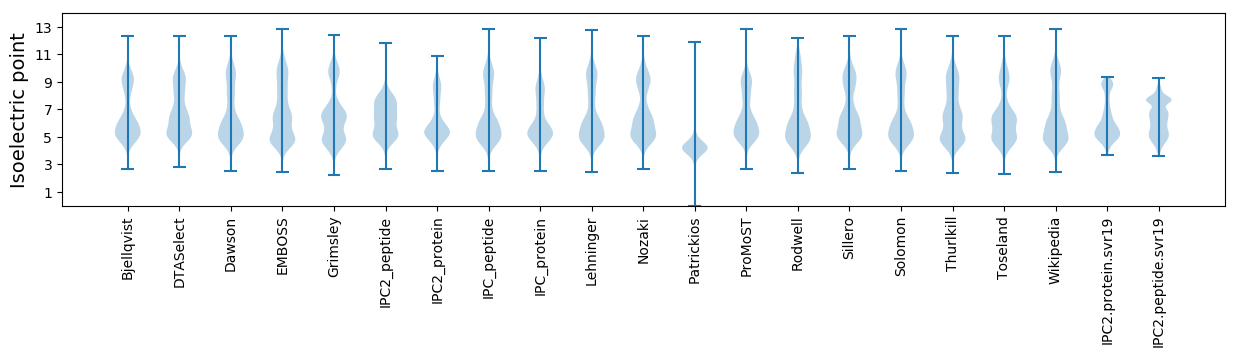
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L0BC87|A0A1L0BC87_9ASCO Protein YIP OS=[Candida] intermedia OX=45354 GN=SAMEA4029010_CIC11G00000003080 PE=3 SV=1
MM1 pKa = 7.42SSKK4 pKa = 9.35PTTPSLKK11 pKa = 10.07SRR13 pKa = 11.84EE14 pKa = 4.04AARR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84LLRR22 pKa = 11.84SSGVYY27 pKa = 10.11KK28 pKa = 10.65ILPQSPKK35 pKa = 8.54IHH37 pKa = 6.3YY38 pKa = 8.79PSEE41 pKa = 4.11VLPLEE46 pKa = 4.32SDD48 pKa = 4.37HH49 pKa = 7.38IQEE52 pKa = 4.5GLRR55 pKa = 11.84ALQEE59 pKa = 4.28SLGEE63 pKa = 4.28LEE65 pKa = 5.81SNMCEE70 pKa = 3.67LAKK73 pKa = 10.34IHH75 pKa = 6.19SAVNSGFNEE84 pKa = 3.73PFAAFLYY91 pKa = 10.7GLLITMFCNNFPGCPTRR108 pKa = 11.84DD109 pKa = 3.27TYY111 pKa = 11.33EE112 pKa = 3.9YY113 pKa = 11.06NQDD116 pKa = 3.21NGQKK120 pKa = 10.15EE121 pKa = 4.37RR122 pKa = 11.84VLLLQKK128 pKa = 10.07RR129 pKa = 11.84VHH131 pKa = 6.43AARR134 pKa = 11.84AEE136 pKa = 3.91NEE138 pKa = 3.96RR139 pKa = 11.84LKK141 pKa = 10.65QQLASQSTSQRR152 pKa = 11.84TIASRR157 pKa = 11.84PALSGQNQAIRR168 pKa = 11.84RR169 pKa = 11.84IGVPSGPSFGNSTRR183 pKa = 11.84GAPLSRR189 pKa = 11.84QKK191 pKa = 10.9KK192 pKa = 8.67VVVAHH197 pKa = 7.56DD198 pKa = 3.94DD199 pKa = 3.9TYY201 pKa = 11.05STTDD205 pKa = 3.16SFVDD209 pKa = 3.35IPTSSKK215 pKa = 10.03LASRR219 pKa = 11.84IQKK222 pKa = 9.08PATAVRR228 pKa = 11.84PGSLNSTGPNLDD240 pKa = 3.09QPPRR244 pKa = 11.84YY245 pKa = 9.17MRR247 pKa = 11.84GLFDD251 pKa = 3.77KK252 pKa = 10.32TSSSNVRR259 pKa = 11.84RR260 pKa = 11.84NLDD263 pKa = 3.18TKK265 pKa = 10.18TRR267 pKa = 11.84ARR269 pKa = 11.84GPTTSSARR277 pKa = 11.84RR278 pKa = 11.84QVEE281 pKa = 3.87RR282 pKa = 11.84ASRR285 pKa = 11.84LAGRR289 pKa = 11.84PPFRR293 pKa = 5.92
MM1 pKa = 7.42SSKK4 pKa = 9.35PTTPSLKK11 pKa = 10.07SRR13 pKa = 11.84EE14 pKa = 4.04AARR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84LLRR22 pKa = 11.84SSGVYY27 pKa = 10.11KK28 pKa = 10.65ILPQSPKK35 pKa = 8.54IHH37 pKa = 6.3YY38 pKa = 8.79PSEE41 pKa = 4.11VLPLEE46 pKa = 4.32SDD48 pKa = 4.37HH49 pKa = 7.38IQEE52 pKa = 4.5GLRR55 pKa = 11.84ALQEE59 pKa = 4.28SLGEE63 pKa = 4.28LEE65 pKa = 5.81SNMCEE70 pKa = 3.67LAKK73 pKa = 10.34IHH75 pKa = 6.19SAVNSGFNEE84 pKa = 3.73PFAAFLYY91 pKa = 10.7GLLITMFCNNFPGCPTRR108 pKa = 11.84DD109 pKa = 3.27TYY111 pKa = 11.33EE112 pKa = 3.9YY113 pKa = 11.06NQDD116 pKa = 3.21NGQKK120 pKa = 10.15EE121 pKa = 4.37RR122 pKa = 11.84VLLLQKK128 pKa = 10.07RR129 pKa = 11.84VHH131 pKa = 6.43AARR134 pKa = 11.84AEE136 pKa = 3.91NEE138 pKa = 3.96RR139 pKa = 11.84LKK141 pKa = 10.65QQLASQSTSQRR152 pKa = 11.84TIASRR157 pKa = 11.84PALSGQNQAIRR168 pKa = 11.84RR169 pKa = 11.84IGVPSGPSFGNSTRR183 pKa = 11.84GAPLSRR189 pKa = 11.84QKK191 pKa = 10.9KK192 pKa = 8.67VVVAHH197 pKa = 7.56DD198 pKa = 3.94DD199 pKa = 3.9TYY201 pKa = 11.05STTDD205 pKa = 3.16SFVDD209 pKa = 3.35IPTSSKK215 pKa = 10.03LASRR219 pKa = 11.84IQKK222 pKa = 9.08PATAVRR228 pKa = 11.84PGSLNSTGPNLDD240 pKa = 3.09QPPRR244 pKa = 11.84YY245 pKa = 9.17MRR247 pKa = 11.84GLFDD251 pKa = 3.77KK252 pKa = 10.32TSSSNVRR259 pKa = 11.84RR260 pKa = 11.84NLDD263 pKa = 3.18TKK265 pKa = 10.18TRR267 pKa = 11.84ARR269 pKa = 11.84GPTTSSARR277 pKa = 11.84RR278 pKa = 11.84QVEE281 pKa = 3.87RR282 pKa = 11.84ASRR285 pKa = 11.84LAGRR289 pKa = 11.84PPFRR293 pKa = 5.92
Molecular weight: 32.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2935372 |
22 |
5583 |
495.8 |
55.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.515 ± 0.04 | 1.121 ± 0.012 |
5.912 ± 0.028 | 6.654 ± 0.041 |
4.419 ± 0.026 | 5.583 ± 0.034 |
2.172 ± 0.018 | 5.723 ± 0.024 |
6.469 ± 0.04 | 10.091 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.055 ± 0.013 | 4.995 ± 0.021 |
4.547 ± 0.028 | 3.784 ± 0.026 |
4.414 ± 0.026 | 8.707 ± 0.07 |
5.894 ± 0.075 | 6.524 ± 0.026 |
1.064 ± 0.011 | 3.357 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
