
Apis mellifera associated microvirus 18
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 7.27
Get precalculated fractions of proteins
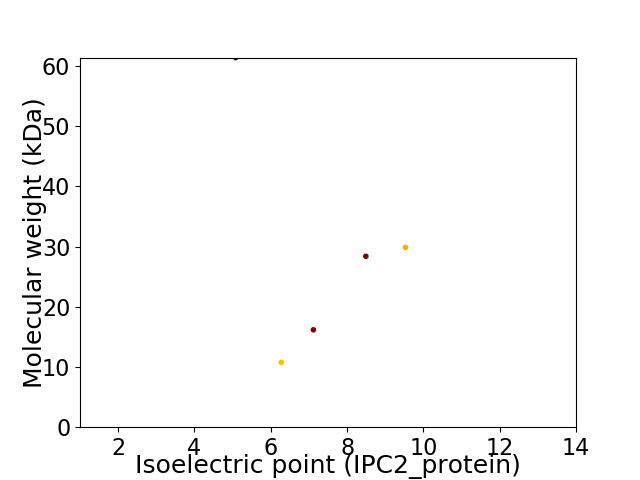
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3S8UUB4|A0A3S8UUB4_9VIRU Replication initiator protein OS=Apis mellifera associated microvirus 18 OX=2494745 PE=4 SV=1
MM1 pKa = 7.18VARR4 pKa = 11.84DD5 pKa = 3.88GDD7 pKa = 3.69PSLRR11 pKa = 11.84EE12 pKa = 4.06DD13 pKa = 4.39LLTMFDD19 pKa = 4.5KK20 pKa = 11.08PFANPRR26 pKa = 11.84LQTSNHH32 pKa = 4.83SFARR36 pKa = 11.84VPQADD41 pKa = 3.5IPRR44 pKa = 11.84STFNRR49 pKa = 11.84SHH51 pKa = 6.84GYY53 pKa = 8.02KK54 pKa = 7.97TTFDD58 pKa = 3.33AGYY61 pKa = 9.42LVPIFVDD68 pKa = 3.68EE69 pKa = 4.73ALPGDD74 pKa = 4.18TFSLHH79 pKa = 4.64MTTFARR85 pKa = 11.84LATPIAPLMDD95 pKa = 4.16NLYY98 pKa = 10.78CDD100 pKa = 3.57VFFFAVPIRR109 pKa = 11.84LVWDD113 pKa = 3.05NWEE116 pKa = 4.44KK117 pKa = 11.05FNGAQDD123 pKa = 4.02NPDD126 pKa = 3.99DD127 pKa = 4.12STDD130 pKa = 3.64FLTPKK135 pKa = 9.55LTTPVGGWATEE146 pKa = 4.0SLSDD150 pKa = 4.5YY151 pKa = 10.44FGLPIGVQHH160 pKa = 6.82ANVHH164 pKa = 5.9AFAHH168 pKa = 6.17RR169 pKa = 11.84AYY171 pKa = 11.1NLIWNEE177 pKa = 3.57WFRR180 pKa = 11.84DD181 pKa = 3.62QNLQDD186 pKa = 3.8SLTVPKK192 pKa = 10.33TDD194 pKa = 4.2GPDD197 pKa = 3.24SATLYY202 pKa = 9.62SLKK205 pKa = 10.4RR206 pKa = 11.84RR207 pKa = 11.84GKK209 pKa = 8.15RR210 pKa = 11.84HH211 pKa = 7.14DD212 pKa = 4.1YY213 pKa = 8.15FTSALPWPQKK223 pKa = 10.69GPAVNLPLGTEE234 pKa = 4.19APITSNFNGAIIGLGEE250 pKa = 4.47ASGQLARR257 pKa = 11.84FRR259 pKa = 11.84PTASTATPAIGAATTSGNPMTASEE283 pKa = 4.06ILEE286 pKa = 4.28WPPSDD291 pKa = 3.41VQYY294 pKa = 11.61YY295 pKa = 10.55NYY297 pKa = 10.91ADD299 pKa = 3.88LSEE302 pKa = 4.4ATAATINQLRR312 pKa = 11.84EE313 pKa = 3.84AFQMQKK319 pKa = 10.54LFEE322 pKa = 5.01RR323 pKa = 11.84DD324 pKa = 3.03ARR326 pKa = 11.84GGTRR330 pKa = 11.84YY331 pKa = 8.58TEE333 pKa = 4.72IIRR336 pKa = 11.84SHH338 pKa = 6.74FGVTSPDD345 pKa = 3.04ARR347 pKa = 11.84LQRR350 pKa = 11.84PEE352 pKa = 3.81YY353 pKa = 10.18LGGGSAPINVAPIPQTSSTDD373 pKa = 2.94ATTPQGNLAAMGTFSHH389 pKa = 6.52SGIGFTKK396 pKa = 10.46SFTEE400 pKa = 3.78HH401 pKa = 7.02CIVIGLISARR411 pKa = 11.84ADD413 pKa = 3.26LNYY416 pKa = 10.22QQGINRR422 pKa = 11.84MWSRR426 pKa = 11.84NTRR429 pKa = 11.84YY430 pKa = 10.59DD431 pKa = 3.87FYY433 pKa = 11.01WPALSHH439 pKa = 6.62IGEE442 pKa = 4.16QAVLNQEE449 pKa = 4.65IYY451 pKa = 10.96AQGTGDD457 pKa = 4.04DD458 pKa = 4.62DD459 pKa = 5.69DD460 pKa = 4.46VFGYY464 pKa = 7.44QEE466 pKa = 4.2RR467 pKa = 11.84YY468 pKa = 9.58AEE470 pKa = 4.04YY471 pKa = 9.71RR472 pKa = 11.84YY473 pKa = 10.38KK474 pKa = 10.77PSLVTGKK481 pKa = 9.86FRR483 pKa = 11.84SDD485 pKa = 2.8ASGSLDD491 pKa = 3.32VWHH494 pKa = 7.26LAQDD498 pKa = 4.08FTGLPVLGDD507 pKa = 3.25TFIQEE512 pKa = 4.55DD513 pKa = 4.42PPVDD517 pKa = 3.27RR518 pKa = 11.84VIAVPSEE525 pKa = 3.67PHH527 pKa = 6.13FLLDD531 pKa = 3.49SAINLKK537 pKa = 9.75CARR540 pKa = 11.84PMPVYY545 pKa = 10.36SVPGLIDD552 pKa = 3.2HH553 pKa = 7.26FF554 pKa = 4.9
MM1 pKa = 7.18VARR4 pKa = 11.84DD5 pKa = 3.88GDD7 pKa = 3.69PSLRR11 pKa = 11.84EE12 pKa = 4.06DD13 pKa = 4.39LLTMFDD19 pKa = 4.5KK20 pKa = 11.08PFANPRR26 pKa = 11.84LQTSNHH32 pKa = 4.83SFARR36 pKa = 11.84VPQADD41 pKa = 3.5IPRR44 pKa = 11.84STFNRR49 pKa = 11.84SHH51 pKa = 6.84GYY53 pKa = 8.02KK54 pKa = 7.97TTFDD58 pKa = 3.33AGYY61 pKa = 9.42LVPIFVDD68 pKa = 3.68EE69 pKa = 4.73ALPGDD74 pKa = 4.18TFSLHH79 pKa = 4.64MTTFARR85 pKa = 11.84LATPIAPLMDD95 pKa = 4.16NLYY98 pKa = 10.78CDD100 pKa = 3.57VFFFAVPIRR109 pKa = 11.84LVWDD113 pKa = 3.05NWEE116 pKa = 4.44KK117 pKa = 11.05FNGAQDD123 pKa = 4.02NPDD126 pKa = 3.99DD127 pKa = 4.12STDD130 pKa = 3.64FLTPKK135 pKa = 9.55LTTPVGGWATEE146 pKa = 4.0SLSDD150 pKa = 4.5YY151 pKa = 10.44FGLPIGVQHH160 pKa = 6.82ANVHH164 pKa = 5.9AFAHH168 pKa = 6.17RR169 pKa = 11.84AYY171 pKa = 11.1NLIWNEE177 pKa = 3.57WFRR180 pKa = 11.84DD181 pKa = 3.62QNLQDD186 pKa = 3.8SLTVPKK192 pKa = 10.33TDD194 pKa = 4.2GPDD197 pKa = 3.24SATLYY202 pKa = 9.62SLKK205 pKa = 10.4RR206 pKa = 11.84RR207 pKa = 11.84GKK209 pKa = 8.15RR210 pKa = 11.84HH211 pKa = 7.14DD212 pKa = 4.1YY213 pKa = 8.15FTSALPWPQKK223 pKa = 10.69GPAVNLPLGTEE234 pKa = 4.19APITSNFNGAIIGLGEE250 pKa = 4.47ASGQLARR257 pKa = 11.84FRR259 pKa = 11.84PTASTATPAIGAATTSGNPMTASEE283 pKa = 4.06ILEE286 pKa = 4.28WPPSDD291 pKa = 3.41VQYY294 pKa = 11.61YY295 pKa = 10.55NYY297 pKa = 10.91ADD299 pKa = 3.88LSEE302 pKa = 4.4ATAATINQLRR312 pKa = 11.84EE313 pKa = 3.84AFQMQKK319 pKa = 10.54LFEE322 pKa = 5.01RR323 pKa = 11.84DD324 pKa = 3.03ARR326 pKa = 11.84GGTRR330 pKa = 11.84YY331 pKa = 8.58TEE333 pKa = 4.72IIRR336 pKa = 11.84SHH338 pKa = 6.74FGVTSPDD345 pKa = 3.04ARR347 pKa = 11.84LQRR350 pKa = 11.84PEE352 pKa = 3.81YY353 pKa = 10.18LGGGSAPINVAPIPQTSSTDD373 pKa = 2.94ATTPQGNLAAMGTFSHH389 pKa = 6.52SGIGFTKK396 pKa = 10.46SFTEE400 pKa = 3.78HH401 pKa = 7.02CIVIGLISARR411 pKa = 11.84ADD413 pKa = 3.26LNYY416 pKa = 10.22QQGINRR422 pKa = 11.84MWSRR426 pKa = 11.84NTRR429 pKa = 11.84YY430 pKa = 10.59DD431 pKa = 3.87FYY433 pKa = 11.01WPALSHH439 pKa = 6.62IGEE442 pKa = 4.16QAVLNQEE449 pKa = 4.65IYY451 pKa = 10.96AQGTGDD457 pKa = 4.04DD458 pKa = 4.62DD459 pKa = 5.69DD460 pKa = 4.46VFGYY464 pKa = 7.44QEE466 pKa = 4.2RR467 pKa = 11.84YY468 pKa = 9.58AEE470 pKa = 4.04YY471 pKa = 9.71RR472 pKa = 11.84YY473 pKa = 10.38KK474 pKa = 10.77PSLVTGKK481 pKa = 9.86FRR483 pKa = 11.84SDD485 pKa = 2.8ASGSLDD491 pKa = 3.32VWHH494 pKa = 7.26LAQDD498 pKa = 4.08FTGLPVLGDD507 pKa = 3.25TFIQEE512 pKa = 4.55DD513 pKa = 4.42PPVDD517 pKa = 3.27RR518 pKa = 11.84VIAVPSEE525 pKa = 3.67PHH527 pKa = 6.13FLLDD531 pKa = 3.49SAINLKK537 pKa = 9.75CARR540 pKa = 11.84PMPVYY545 pKa = 10.36SVPGLIDD552 pKa = 3.2HH553 pKa = 7.26FF554 pKa = 4.9
Molecular weight: 61.37 kDa
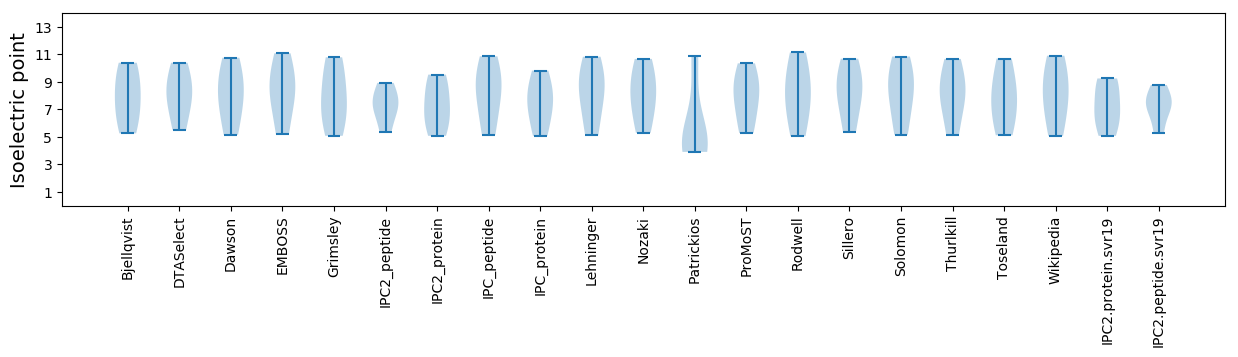
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3S8UU77|A0A3S8UU77_9VIRU Nonstructural protein OS=Apis mellifera associated microvirus 18 OX=2494745 PE=4 SV=1
MM1 pKa = 7.99RR2 pKa = 11.84PPDD5 pKa = 3.76ARR7 pKa = 11.84VLCARR12 pKa = 11.84SHH14 pKa = 6.69RR15 pKa = 11.84SLLSVAMWKK24 pKa = 10.55LIFPVLRR31 pKa = 11.84ASIRR35 pKa = 11.84SIDD38 pKa = 4.31PIVGGALIGGGFNLLSGLFGGSSSLKK64 pKa = 9.63AQKK67 pKa = 10.08AANKK71 pKa = 9.88ANLKK75 pKa = 9.35IAHH78 pKa = 7.02DD79 pKa = 3.4NRR81 pKa = 11.84VFQEE85 pKa = 4.19RR86 pKa = 11.84MSSTAHH92 pKa = 4.67QRR94 pKa = 11.84QVEE97 pKa = 4.25DD98 pKa = 3.34MRR100 pKa = 11.84AAGLNPILSATQGGSSAPQGSSATMVAEE128 pKa = 4.34DD129 pKa = 4.27AMGQAVSDD137 pKa = 3.6AGHH140 pKa = 6.42RR141 pKa = 11.84VASTARR147 pKa = 11.84DD148 pKa = 3.64MFRR151 pKa = 11.84LQTDD155 pKa = 3.47VALADD160 pKa = 3.77AEE162 pKa = 4.39IAGKK166 pKa = 9.9KK167 pKa = 9.23AAALNAASQAEE178 pKa = 4.58LNLATAKK185 pKa = 10.54NVTLQRR191 pKa = 11.84PKK193 pKa = 10.84LIAEE197 pKa = 4.62GRR199 pKa = 11.84ASGHH203 pKa = 6.01SADD206 pKa = 3.75ATIGEE211 pKa = 4.27AAVRR215 pKa = 11.84KK216 pKa = 9.96HH217 pKa = 4.76MSEE220 pKa = 3.89YY221 pKa = 10.63DD222 pKa = 3.66KK223 pKa = 11.62KK224 pKa = 10.6FVPYY228 pKa = 10.06DD229 pKa = 3.54AVSSRR234 pKa = 11.84IFDD237 pKa = 4.99LIGNTGKK244 pKa = 10.57AVGTAIHH251 pKa = 7.34GIRR254 pKa = 11.84NGFSGSKK261 pKa = 8.75NQPRR265 pKa = 11.84STPKK269 pKa = 9.92PPFNPRR275 pKa = 11.84YY276 pKa = 9.54PMGGKK281 pKa = 9.37SGRR284 pKa = 3.59
MM1 pKa = 7.99RR2 pKa = 11.84PPDD5 pKa = 3.76ARR7 pKa = 11.84VLCARR12 pKa = 11.84SHH14 pKa = 6.69RR15 pKa = 11.84SLLSVAMWKK24 pKa = 10.55LIFPVLRR31 pKa = 11.84ASIRR35 pKa = 11.84SIDD38 pKa = 4.31PIVGGALIGGGFNLLSGLFGGSSSLKK64 pKa = 9.63AQKK67 pKa = 10.08AANKK71 pKa = 9.88ANLKK75 pKa = 9.35IAHH78 pKa = 7.02DD79 pKa = 3.4NRR81 pKa = 11.84VFQEE85 pKa = 4.19RR86 pKa = 11.84MSSTAHH92 pKa = 4.67QRR94 pKa = 11.84QVEE97 pKa = 4.25DD98 pKa = 3.34MRR100 pKa = 11.84AAGLNPILSATQGGSSAPQGSSATMVAEE128 pKa = 4.34DD129 pKa = 4.27AMGQAVSDD137 pKa = 3.6AGHH140 pKa = 6.42RR141 pKa = 11.84VASTARR147 pKa = 11.84DD148 pKa = 3.64MFRR151 pKa = 11.84LQTDD155 pKa = 3.47VALADD160 pKa = 3.77AEE162 pKa = 4.39IAGKK166 pKa = 9.9KK167 pKa = 9.23AAALNAASQAEE178 pKa = 4.58LNLATAKK185 pKa = 10.54NVTLQRR191 pKa = 11.84PKK193 pKa = 10.84LIAEE197 pKa = 4.62GRR199 pKa = 11.84ASGHH203 pKa = 6.01SADD206 pKa = 3.75ATIGEE211 pKa = 4.27AAVRR215 pKa = 11.84KK216 pKa = 9.96HH217 pKa = 4.76MSEE220 pKa = 3.89YY221 pKa = 10.63DD222 pKa = 3.66KK223 pKa = 11.62KK224 pKa = 10.6FVPYY228 pKa = 10.06DD229 pKa = 3.54AVSSRR234 pKa = 11.84IFDD237 pKa = 4.99LIGNTGKK244 pKa = 10.57AVGTAIHH251 pKa = 7.34GIRR254 pKa = 11.84NGFSGSKK261 pKa = 8.75NQPRR265 pKa = 11.84STPKK269 pKa = 9.92PPFNPRR275 pKa = 11.84YY276 pKa = 9.54PMGGKK281 pKa = 9.37SGRR284 pKa = 3.59
Molecular weight: 29.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1323 |
96 |
554 |
264.6 |
29.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.582 ± 1.435 | 0.907 ± 0.337 |
6.274 ± 0.596 | 4.989 ± 0.97 |
4.837 ± 0.602 | 6.878 ± 0.793 |
2.872 ± 0.449 | 4.157 ± 0.476 |
4.837 ± 1.253 | 7.559 ± 0.308 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.041 ± 0.388 | 3.704 ± 0.446 |
6.803 ± 0.686 | 3.855 ± 0.205 |
7.029 ± 0.774 | 7.483 ± 0.626 |
5.745 ± 1.016 | 5.14 ± 0.294 |
1.134 ± 0.347 | 3.175 ± 0.672 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
