
Miniopterus associated gemycircularvirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus minio1
Average proteome isoelectric point is 5.56
Get precalculated fractions of proteins
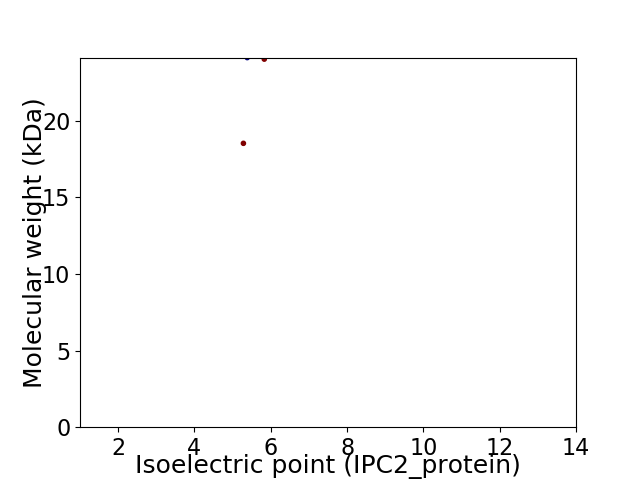
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0D3MDS0|A0A0D3MDS0_9VIRU Replication-associated protein OS=Miniopterus associated gemycircularvirus 1 OX=1985387 PE=4 SV=1
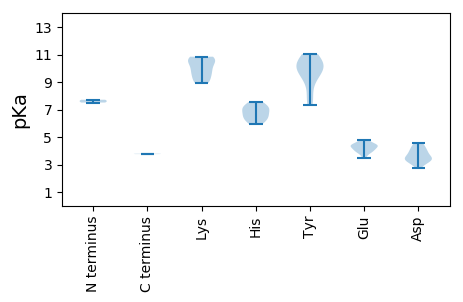
MM1 pKa = 7.51GCLGGASLGVGMQAHH16 pKa = 7.1LSAPLGAGAAAHH28 pKa = 6.69GGPTSTLVDD37 pKa = 4.42FLTEE41 pKa = 3.91RR42 pKa = 11.84PMSLVLWGPTRR53 pKa = 11.84TGKK56 pKa = 9.03TSWARR61 pKa = 11.84SLGPHH66 pKa = 7.54AYY68 pKa = 9.16TMGMVSGNKK77 pKa = 9.57LLADD81 pKa = 3.91MEE83 pKa = 4.45DD84 pKa = 3.16AEE86 pKa = 4.63YY87 pKa = 11.04AVFDD91 pKa = 4.56DD92 pKa = 4.22MRR94 pKa = 11.84GGMKK98 pKa = 9.81FFPAWKK104 pKa = 9.19EE105 pKa = 3.46WFGAQKK111 pKa = 10.67VVTIKK116 pKa = 10.66CLYY119 pKa = 10.39KK120 pKa = 10.63EE121 pKa = 4.1PQQVNWGKK129 pKa = 9.84PIIWISNKK137 pKa = 9.96DD138 pKa = 3.44PRR140 pKa = 11.84EE141 pKa = 4.11DD142 pKa = 3.05MDD144 pKa = 3.86MGDD147 pKa = 4.04TEE149 pKa = 4.37WLEE152 pKa = 4.33GNASIVYY159 pKa = 10.25VGDD162 pKa = 3.74SLISRR167 pKa = 11.84ASTEE171 pKa = 3.76
MM1 pKa = 7.51GCLGGASLGVGMQAHH16 pKa = 7.1LSAPLGAGAAAHH28 pKa = 6.69GGPTSTLVDD37 pKa = 4.42FLTEE41 pKa = 3.91RR42 pKa = 11.84PMSLVLWGPTRR53 pKa = 11.84TGKK56 pKa = 9.03TSWARR61 pKa = 11.84SLGPHH66 pKa = 7.54AYY68 pKa = 9.16TMGMVSGNKK77 pKa = 9.57LLADD81 pKa = 3.91MEE83 pKa = 4.45DD84 pKa = 3.16AEE86 pKa = 4.63YY87 pKa = 11.04AVFDD91 pKa = 4.56DD92 pKa = 4.22MRR94 pKa = 11.84GGMKK98 pKa = 9.81FFPAWKK104 pKa = 9.19EE105 pKa = 3.46WFGAQKK111 pKa = 10.67VVTIKK116 pKa = 10.66CLYY119 pKa = 10.39KK120 pKa = 10.63EE121 pKa = 4.1PQQVNWGKK129 pKa = 9.84PIIWISNKK137 pKa = 9.96DD138 pKa = 3.44PRR140 pKa = 11.84EE141 pKa = 4.11DD142 pKa = 3.05MDD144 pKa = 3.86MGDD147 pKa = 4.04TEE149 pKa = 4.37WLEE152 pKa = 4.33GNASIVYY159 pKa = 10.25VGDD162 pKa = 3.74SLISRR167 pKa = 11.84ASTEE171 pKa = 3.76
Molecular weight: 18.53 kDa
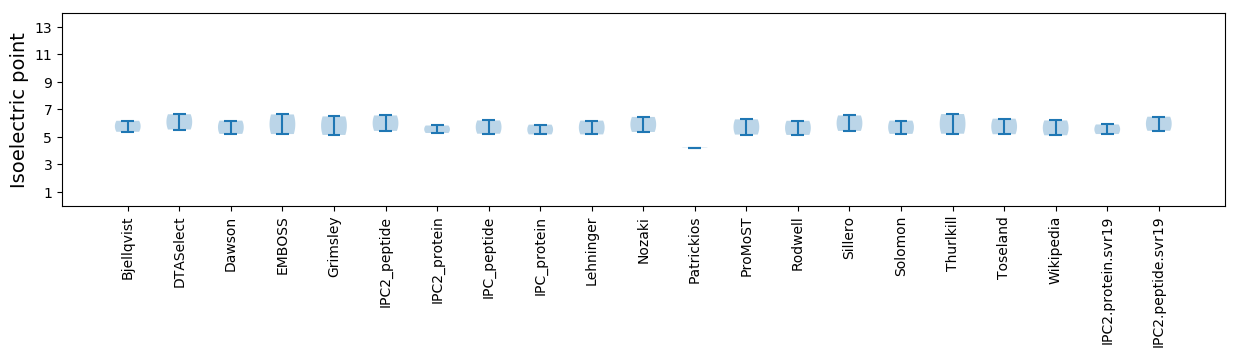
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D3MDS0|A0A0D3MDS0_9VIRU Replication-associated protein OS=Miniopterus associated gemycircularvirus 1 OX=1985387 PE=4 SV=1
MM1 pKa = 7.71PFKK4 pKa = 10.55FQSRR8 pKa = 11.84YY9 pKa = 9.78VLLTYY14 pKa = 8.03SQCGEE19 pKa = 4.37LDD21 pKa = 3.4PFDD24 pKa = 4.25VVNHH28 pKa = 6.39LSSLGAEE35 pKa = 4.35CIVGRR40 pKa = 11.84EE41 pKa = 3.92PHH43 pKa = 6.86ADD45 pKa = 3.28GGTHH49 pKa = 6.2LHH51 pKa = 6.48AFVDD55 pKa = 4.57FGRR58 pKa = 11.84RR59 pKa = 11.84YY60 pKa = 7.34STRR63 pKa = 11.84RR64 pKa = 11.84SDD66 pKa = 3.36AFDD69 pKa = 3.04VGGYY73 pKa = 9.35HH74 pKa = 7.24PNIAPSRR81 pKa = 11.84GRR83 pKa = 11.84AEE85 pKa = 4.5GGWDD89 pKa = 3.43YY90 pKa = 10.96ATKK93 pKa = 10.59EE94 pKa = 3.88GDD96 pKa = 3.21IVAGGLEE103 pKa = 4.11RR104 pKa = 11.84PSRR107 pKa = 11.84SRR109 pKa = 11.84VSRR112 pKa = 11.84NDD114 pKa = 4.08HH115 pKa = 5.97IWDD118 pKa = 3.7EE119 pKa = 4.77VINQPDD125 pKa = 3.43EE126 pKa = 4.19SAFWEE131 pKa = 4.8CVQRR135 pKa = 11.84LDD137 pKa = 3.9PKK139 pKa = 10.84AAATQFTQLRR149 pKa = 11.84AYY151 pKa = 10.2ASWAYY156 pKa = 9.98AVQPEE161 pKa = 4.39QYY163 pKa = 10.03EE164 pKa = 4.4PNPEE168 pKa = 4.03HH169 pKa = 7.15AFDD172 pKa = 3.46TGMVPEE178 pKa = 4.42LDD180 pKa = 2.75RR181 pKa = 11.84WRR183 pKa = 11.84RR184 pKa = 11.84EE185 pKa = 3.71SLGDD189 pKa = 3.55DD190 pKa = 3.44RR191 pKa = 11.84QGTFAFSRR199 pKa = 11.84SGRR202 pKa = 11.84KK203 pKa = 8.94RR204 pKa = 11.84PALYY208 pKa = 10.4RR209 pKa = 11.84NRR211 pKa = 3.78
MM1 pKa = 7.71PFKK4 pKa = 10.55FQSRR8 pKa = 11.84YY9 pKa = 9.78VLLTYY14 pKa = 8.03SQCGEE19 pKa = 4.37LDD21 pKa = 3.4PFDD24 pKa = 4.25VVNHH28 pKa = 6.39LSSLGAEE35 pKa = 4.35CIVGRR40 pKa = 11.84EE41 pKa = 3.92PHH43 pKa = 6.86ADD45 pKa = 3.28GGTHH49 pKa = 6.2LHH51 pKa = 6.48AFVDD55 pKa = 4.57FGRR58 pKa = 11.84RR59 pKa = 11.84YY60 pKa = 7.34STRR63 pKa = 11.84RR64 pKa = 11.84SDD66 pKa = 3.36AFDD69 pKa = 3.04VGGYY73 pKa = 9.35HH74 pKa = 7.24PNIAPSRR81 pKa = 11.84GRR83 pKa = 11.84AEE85 pKa = 4.5GGWDD89 pKa = 3.43YY90 pKa = 10.96ATKK93 pKa = 10.59EE94 pKa = 3.88GDD96 pKa = 3.21IVAGGLEE103 pKa = 4.11RR104 pKa = 11.84PSRR107 pKa = 11.84SRR109 pKa = 11.84VSRR112 pKa = 11.84NDD114 pKa = 4.08HH115 pKa = 5.97IWDD118 pKa = 3.7EE119 pKa = 4.77VINQPDD125 pKa = 3.43EE126 pKa = 4.19SAFWEE131 pKa = 4.8CVQRR135 pKa = 11.84LDD137 pKa = 3.9PKK139 pKa = 10.84AAATQFTQLRR149 pKa = 11.84AYY151 pKa = 10.2ASWAYY156 pKa = 9.98AVQPEE161 pKa = 4.39QYY163 pKa = 10.03EE164 pKa = 4.4PNPEE168 pKa = 4.03HH169 pKa = 7.15AFDD172 pKa = 3.46TGMVPEE178 pKa = 4.42LDD180 pKa = 2.75RR181 pKa = 11.84WRR183 pKa = 11.84RR184 pKa = 11.84EE185 pKa = 3.71SLGDD189 pKa = 3.55DD190 pKa = 3.44RR191 pKa = 11.84QGTFAFSRR199 pKa = 11.84SGRR202 pKa = 11.84KK203 pKa = 8.94RR204 pKa = 11.84PALYY208 pKa = 10.4RR209 pKa = 11.84NRR211 pKa = 3.78
Molecular weight: 24.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
382 |
171 |
211 |
191.0 |
21.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.162 ± 0.113 | 1.309 ± 0.081 |
6.806 ± 0.558 | 6.021 ± 0.441 |
4.188 ± 0.736 | 10.209 ± 1.206 |
2.618 ± 0.503 | 2.88 ± 0.366 |
3.403 ± 1.083 | 6.806 ± 0.804 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.141 ± 1.576 | 2.618 ± 0.162 |
5.759 ± 0.289 | 3.403 ± 0.62 |
7.592 ± 2.377 | 7.068 ± 0.029 |
4.712 ± 0.661 | 5.759 ± 0.052 |
3.141 ± 0.554 | 3.403 ± 0.62 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
