
Wuhan Insect virus 4
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Betarhabdovirinae; Cytorhabdovirus; Wuhan 4 insect cytorhabdovirus
Average proteome isoelectric point is 6.74
Get precalculated fractions of proteins
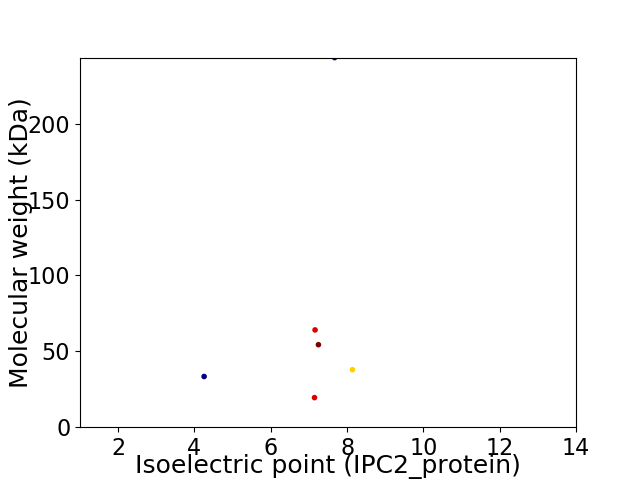
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
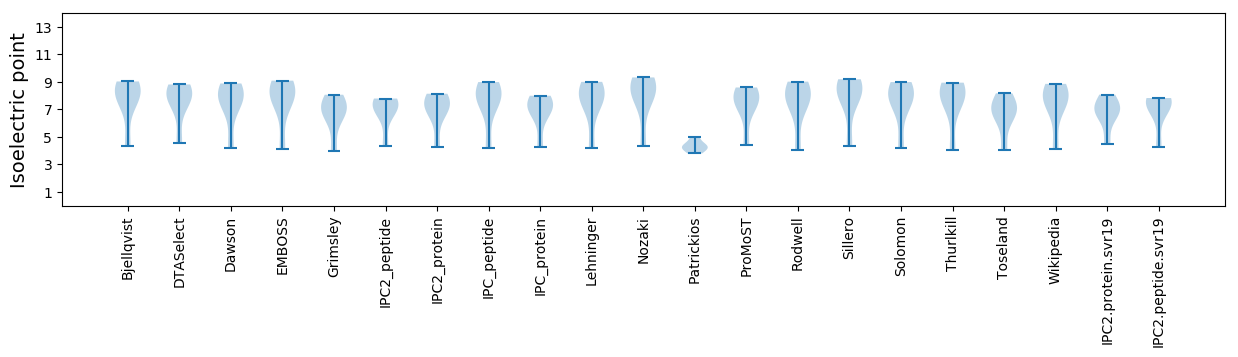
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B5KRN7|A0A0B5KRN7_9RHAB 4b protein OS=Wuhan Insect virus 4 OX=1608109 GN=4b PE=4 SV=1
MM1 pKa = 7.64AKK3 pKa = 9.97SQGFDD8 pKa = 3.26DD9 pKa = 4.89LAFGMSPHH17 pKa = 6.84HH18 pKa = 6.91NPFNDD23 pKa = 3.76DD24 pKa = 3.69ADD26 pKa = 3.92SDD28 pKa = 4.21EE29 pKa = 4.88YY30 pKa = 11.36NDD32 pKa = 4.54NVTQVPSPTVEE43 pKa = 4.62DD44 pKa = 3.84SDD46 pKa = 4.67AADD49 pKa = 3.88PSDD52 pKa = 3.75YY53 pKa = 10.43QSKK56 pKa = 10.95KK57 pKa = 10.9DD58 pKa = 3.25IDD60 pKa = 4.25QIILDD65 pKa = 4.34LNEE68 pKa = 4.05SCAEE72 pKa = 4.3LGVQTDD78 pKa = 3.47SEE80 pKa = 4.66MINAVLAMSEE90 pKa = 4.65SYY92 pKa = 11.17TMDD95 pKa = 3.26SFSLRR100 pKa = 11.84WFAFGMTYY108 pKa = 11.2ANNQRR113 pKa = 11.84IIPQLSGVLGDD124 pKa = 4.47LRR126 pKa = 11.84AEE128 pKa = 4.2IKK130 pKa = 10.58ALQQTNSSISKK141 pKa = 8.51TSSDD145 pKa = 2.89ITKK148 pKa = 10.7KK149 pKa = 9.72MVSVKK154 pKa = 10.73NDD156 pKa = 3.18VLDD159 pKa = 4.1GFEE162 pKa = 4.18KK163 pKa = 10.76LRR165 pKa = 11.84ANVLDD170 pKa = 3.95KK171 pKa = 11.29VVEE174 pKa = 4.4TSAHH178 pKa = 4.82IMARR182 pKa = 11.84DD183 pKa = 3.7EE184 pKa = 4.33NKK186 pKa = 10.29EE187 pKa = 3.84ATTQNSPSIYY197 pKa = 9.22PKK199 pKa = 9.97KK200 pKa = 10.71DD201 pKa = 3.23KK202 pKa = 10.94IEE204 pKa = 4.09LAGPTTIADD213 pKa = 3.79SQSVVMSSSPIFNDD227 pKa = 3.24PVEE230 pKa = 4.36PKK232 pKa = 9.52MVNPLLKK239 pKa = 10.21EE240 pKa = 3.81KK241 pKa = 10.72RR242 pKa = 11.84NLLIEE247 pKa = 4.4SGADD251 pKa = 3.1KK252 pKa = 10.96KK253 pKa = 10.91KK254 pKa = 10.67LRR256 pKa = 11.84EE257 pKa = 4.08LTDD260 pKa = 4.14DD261 pKa = 4.4EE262 pKa = 5.67IEE264 pKa = 4.2LLVIDD269 pKa = 4.98EE270 pKa = 4.54EE271 pKa = 5.18LEE273 pKa = 4.15LSQDD277 pKa = 3.39PTKK280 pKa = 10.75QEE282 pKa = 4.05SADD285 pKa = 3.42QLMAVLMSRR294 pKa = 11.84LLDD297 pKa = 3.53MDD299 pKa = 4.32LMM301 pKa = 5.99
MM1 pKa = 7.64AKK3 pKa = 9.97SQGFDD8 pKa = 3.26DD9 pKa = 4.89LAFGMSPHH17 pKa = 6.84HH18 pKa = 6.91NPFNDD23 pKa = 3.76DD24 pKa = 3.69ADD26 pKa = 3.92SDD28 pKa = 4.21EE29 pKa = 4.88YY30 pKa = 11.36NDD32 pKa = 4.54NVTQVPSPTVEE43 pKa = 4.62DD44 pKa = 3.84SDD46 pKa = 4.67AADD49 pKa = 3.88PSDD52 pKa = 3.75YY53 pKa = 10.43QSKK56 pKa = 10.95KK57 pKa = 10.9DD58 pKa = 3.25IDD60 pKa = 4.25QIILDD65 pKa = 4.34LNEE68 pKa = 4.05SCAEE72 pKa = 4.3LGVQTDD78 pKa = 3.47SEE80 pKa = 4.66MINAVLAMSEE90 pKa = 4.65SYY92 pKa = 11.17TMDD95 pKa = 3.26SFSLRR100 pKa = 11.84WFAFGMTYY108 pKa = 11.2ANNQRR113 pKa = 11.84IIPQLSGVLGDD124 pKa = 4.47LRR126 pKa = 11.84AEE128 pKa = 4.2IKK130 pKa = 10.58ALQQTNSSISKK141 pKa = 8.51TSSDD145 pKa = 2.89ITKK148 pKa = 10.7KK149 pKa = 9.72MVSVKK154 pKa = 10.73NDD156 pKa = 3.18VLDD159 pKa = 4.1GFEE162 pKa = 4.18KK163 pKa = 10.76LRR165 pKa = 11.84ANVLDD170 pKa = 3.95KK171 pKa = 11.29VVEE174 pKa = 4.4TSAHH178 pKa = 4.82IMARR182 pKa = 11.84DD183 pKa = 3.7EE184 pKa = 4.33NKK186 pKa = 10.29EE187 pKa = 3.84ATTQNSPSIYY197 pKa = 9.22PKK199 pKa = 9.97KK200 pKa = 10.71DD201 pKa = 3.23KK202 pKa = 10.94IEE204 pKa = 4.09LAGPTTIADD213 pKa = 3.79SQSVVMSSSPIFNDD227 pKa = 3.24PVEE230 pKa = 4.36PKK232 pKa = 9.52MVNPLLKK239 pKa = 10.21EE240 pKa = 3.81KK241 pKa = 10.72RR242 pKa = 11.84NLLIEE247 pKa = 4.4SGADD251 pKa = 3.1KK252 pKa = 10.96KK253 pKa = 10.91KK254 pKa = 10.67LRR256 pKa = 11.84EE257 pKa = 4.08LTDD260 pKa = 4.14DD261 pKa = 4.4EE262 pKa = 5.67IEE264 pKa = 4.2LLVIDD269 pKa = 4.98EE270 pKa = 4.54EE271 pKa = 5.18LEE273 pKa = 4.15LSQDD277 pKa = 3.39PTKK280 pKa = 10.75QEE282 pKa = 4.05SADD285 pKa = 3.42QLMAVLMSRR294 pKa = 11.84LLDD297 pKa = 3.53MDD299 pKa = 4.32LMM301 pKa = 5.99
Molecular weight: 33.41 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B5KTL5|A0A0B5KTL5_9RHAB Putative matrix protein OS=Wuhan Insect virus 4 OX=1608109 GN=ORF4 PE=4 SV=1
MM1 pKa = 7.32SRR3 pKa = 11.84SGLISVNDD11 pKa = 3.82AKK13 pKa = 11.11AVVAINGSLTSFIDD27 pKa = 3.25SSKK30 pKa = 9.92VYY32 pKa = 9.99EE33 pKa = 4.23GRR35 pKa = 11.84ARR37 pKa = 11.84HH38 pKa = 5.65IARR41 pKa = 11.84KK42 pKa = 9.84RR43 pKa = 11.84EE44 pKa = 3.94VNVKK48 pKa = 9.79ILATKK53 pKa = 9.73DD54 pKa = 3.07NKK56 pKa = 10.94YY57 pKa = 11.17LMRR60 pKa = 11.84CFPLLDD66 pKa = 4.09NNDD69 pKa = 3.59VSEE72 pKa = 4.08MRR74 pKa = 11.84ADD76 pKa = 3.52KK77 pKa = 11.08KK78 pKa = 9.98NNKK81 pKa = 8.61YY82 pKa = 10.09IHH84 pKa = 6.81IGCISVSIEE93 pKa = 3.77PLIHH97 pKa = 6.72ARR99 pKa = 11.84YY100 pKa = 8.2LKK102 pKa = 10.57KK103 pKa = 10.52YY104 pKa = 7.94GKK106 pKa = 10.07EE107 pKa = 3.79MEE109 pKa = 4.86GICAIMDD116 pKa = 3.9TTFHH120 pKa = 7.58DD121 pKa = 3.68IDD123 pKa = 3.52QSIISLHH130 pKa = 6.49RR131 pKa = 11.84FDD133 pKa = 5.94LSHH136 pKa = 6.73KK137 pKa = 10.0RR138 pKa = 11.84ADD140 pKa = 5.27FICQPNHH147 pKa = 6.35SLSITDD153 pKa = 3.85DD154 pKa = 3.23NLMRR158 pKa = 11.84RR159 pKa = 11.84ISVLVIFDD167 pKa = 3.95CPKK170 pKa = 9.56VDD172 pKa = 4.24EE173 pKa = 4.74GNEE176 pKa = 3.97LFNICIGHH184 pKa = 6.43ITTCTNTLNPTSLEE198 pKa = 4.22GEE200 pKa = 4.24TSFGVIGSTPVEE212 pKa = 4.14FAAVKK217 pKa = 10.33DD218 pKa = 4.18HH219 pKa = 7.32IEE221 pKa = 3.99NALNPMGGTPCIEE234 pKa = 4.15LSEE237 pKa = 5.55DD238 pKa = 3.65GADD241 pKa = 4.61DD242 pKa = 4.95IIMKK246 pKa = 10.3KK247 pKa = 10.29KK248 pKa = 10.63SLLSKK253 pKa = 10.54VGLIKK258 pKa = 10.59APKK261 pKa = 8.83VVIRR265 pKa = 11.84KK266 pKa = 8.99NYY268 pKa = 8.83AMRR271 pKa = 11.84PRR273 pKa = 11.84EE274 pKa = 3.74HH275 pKa = 7.24DD276 pKa = 4.24RR277 pKa = 11.84IEE279 pKa = 4.22SSSDD283 pKa = 3.12LKK285 pKa = 10.93RR286 pKa = 11.84CSSLPRR292 pKa = 11.84NSLDD296 pKa = 3.46LAVARR301 pKa = 11.84VAKK304 pKa = 10.47EE305 pKa = 4.06GMFSAAFKK313 pKa = 9.33SHH315 pKa = 7.0RR316 pKa = 11.84LGGDD320 pKa = 2.66IDD322 pKa = 3.71KK323 pKa = 11.35QMILEE328 pKa = 4.21KK329 pKa = 10.49KK330 pKa = 10.26KK331 pKa = 10.64SIVRR335 pKa = 11.84NRR337 pKa = 11.84QNN339 pKa = 2.72
MM1 pKa = 7.32SRR3 pKa = 11.84SGLISVNDD11 pKa = 3.82AKK13 pKa = 11.11AVVAINGSLTSFIDD27 pKa = 3.25SSKK30 pKa = 9.92VYY32 pKa = 9.99EE33 pKa = 4.23GRR35 pKa = 11.84ARR37 pKa = 11.84HH38 pKa = 5.65IARR41 pKa = 11.84KK42 pKa = 9.84RR43 pKa = 11.84EE44 pKa = 3.94VNVKK48 pKa = 9.79ILATKK53 pKa = 9.73DD54 pKa = 3.07NKK56 pKa = 10.94YY57 pKa = 11.17LMRR60 pKa = 11.84CFPLLDD66 pKa = 4.09NNDD69 pKa = 3.59VSEE72 pKa = 4.08MRR74 pKa = 11.84ADD76 pKa = 3.52KK77 pKa = 11.08KK78 pKa = 9.98NNKK81 pKa = 8.61YY82 pKa = 10.09IHH84 pKa = 6.81IGCISVSIEE93 pKa = 3.77PLIHH97 pKa = 6.72ARR99 pKa = 11.84YY100 pKa = 8.2LKK102 pKa = 10.57KK103 pKa = 10.52YY104 pKa = 7.94GKK106 pKa = 10.07EE107 pKa = 3.79MEE109 pKa = 4.86GICAIMDD116 pKa = 3.9TTFHH120 pKa = 7.58DD121 pKa = 3.68IDD123 pKa = 3.52QSIISLHH130 pKa = 6.49RR131 pKa = 11.84FDD133 pKa = 5.94LSHH136 pKa = 6.73KK137 pKa = 10.0RR138 pKa = 11.84ADD140 pKa = 5.27FICQPNHH147 pKa = 6.35SLSITDD153 pKa = 3.85DD154 pKa = 3.23NLMRR158 pKa = 11.84RR159 pKa = 11.84ISVLVIFDD167 pKa = 3.95CPKK170 pKa = 9.56VDD172 pKa = 4.24EE173 pKa = 4.74GNEE176 pKa = 3.97LFNICIGHH184 pKa = 6.43ITTCTNTLNPTSLEE198 pKa = 4.22GEE200 pKa = 4.24TSFGVIGSTPVEE212 pKa = 4.14FAAVKK217 pKa = 10.33DD218 pKa = 4.18HH219 pKa = 7.32IEE221 pKa = 3.99NALNPMGGTPCIEE234 pKa = 4.15LSEE237 pKa = 5.55DD238 pKa = 3.65GADD241 pKa = 4.61DD242 pKa = 4.95IIMKK246 pKa = 10.3KK247 pKa = 10.29KK248 pKa = 10.63SLLSKK253 pKa = 10.54VGLIKK258 pKa = 10.59APKK261 pKa = 8.83VVIRR265 pKa = 11.84KK266 pKa = 8.99NYY268 pKa = 8.83AMRR271 pKa = 11.84PRR273 pKa = 11.84EE274 pKa = 3.74HH275 pKa = 7.24DD276 pKa = 4.24RR277 pKa = 11.84IEE279 pKa = 4.22SSSDD283 pKa = 3.12LKK285 pKa = 10.93RR286 pKa = 11.84CSSLPRR292 pKa = 11.84NSLDD296 pKa = 3.46LAVARR301 pKa = 11.84VAKK304 pKa = 10.47EE305 pKa = 4.06GMFSAAFKK313 pKa = 9.33SHH315 pKa = 7.0RR316 pKa = 11.84LGGDD320 pKa = 2.66IDD322 pKa = 3.71KK323 pKa = 11.35QMILEE328 pKa = 4.21KK329 pKa = 10.49KK330 pKa = 10.26KK331 pKa = 10.64SIVRR335 pKa = 11.84NRR337 pKa = 11.84QNN339 pKa = 2.72
Molecular weight: 37.9 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3973 |
172 |
2105 |
662.2 |
75.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.757 ± 1.186 | 1.988 ± 0.417 |
6.343 ± 0.555 | 5.915 ± 0.408 |
3.398 ± 0.571 | 4.782 ± 0.367 |
2.391 ± 0.367 | 7.501 ± 0.489 |
7.375 ± 0.628 | 9.086 ± 0.336 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.876 ± 0.333 | 5.311 ± 0.259 |
3.574 ± 0.2 | 2.693 ± 0.468 |
5.135 ± 0.649 | 9.665 ± 0.62 |
5.059 ± 0.491 | 5.915 ± 0.128 |
1.309 ± 0.284 | 3.927 ± 0.432 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
