
Tuhoko virus 3
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Rubulavirinae; Pararubulavirus; Tuhoko pararubulavirus 3
Average proteome isoelectric point is 7.04
Get precalculated fractions of proteins
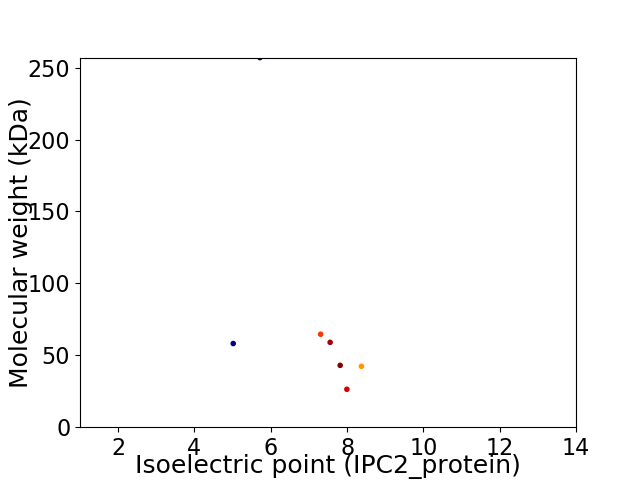
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D8WJ37|D8WJ37_9MONO Phosphoprotein OS=Tuhoko virus 3 OX=798074 GN=V/P PE=3 SV=1
MM1 pKa = 7.63ASLFRR6 pKa = 11.84ALEE9 pKa = 4.01QFTLEE14 pKa = 3.83QDD16 pKa = 2.79QGGRR20 pKa = 11.84GNVGDD25 pKa = 4.39QPPEE29 pKa = 3.9TLQATIKK36 pKa = 10.49VFVINTPDD44 pKa = 2.96PRR46 pKa = 11.84LRR48 pKa = 11.84YY49 pKa = 10.36RR50 pKa = 11.84MMCFCLRR57 pKa = 11.84LIVSNSARR65 pKa = 11.84TGQRR69 pKa = 11.84HH70 pKa = 5.2GALMTLLSLPTAVMQNHH87 pKa = 5.7FRR89 pKa = 11.84TAEE92 pKa = 3.95RR93 pKa = 11.84SPDD96 pKa = 3.6CQIEE100 pKa = 4.28RR101 pKa = 11.84IEE103 pKa = 4.03VDD105 pKa = 3.04GFEE108 pKa = 4.54PGTYY112 pKa = 9.68RR113 pKa = 11.84IRR115 pKa = 11.84TNARR119 pKa = 11.84TPLTHH124 pKa = 6.63GEE126 pKa = 4.27VVALEE131 pKa = 4.17EE132 pKa = 4.17MADD135 pKa = 4.56DD136 pKa = 4.66IPEE139 pKa = 4.17ALANHH144 pKa = 6.27TPFVDD149 pKa = 3.4AQTEE153 pKa = 4.25LEE155 pKa = 4.45EE156 pKa = 5.24CDD158 pKa = 3.62EE159 pKa = 4.19MEE161 pKa = 4.74KK162 pKa = 10.56FLEE165 pKa = 5.01AIYY168 pKa = 9.24STLIQVWIMVTKK180 pKa = 10.91SMTAFDD186 pKa = 4.14QPTGSDD192 pKa = 3.06EE193 pKa = 4.23RR194 pKa = 11.84RR195 pKa = 11.84VAKK198 pKa = 9.97YY199 pKa = 8.5QQQGRR204 pKa = 11.84LNPHH208 pKa = 6.3YY209 pKa = 10.19LLQGEE214 pKa = 4.44VRR216 pKa = 11.84RR217 pKa = 11.84RR218 pKa = 11.84IQLCIRR224 pKa = 11.84QSLPIRR230 pKa = 11.84QFLVNEE236 pKa = 4.35LQTASNQGPITGKK249 pKa = 10.1YY250 pKa = 7.77YY251 pKa = 11.55AMVADD256 pKa = 3.46IGKK259 pKa = 10.03YY260 pKa = 9.88IMNAGMGGFFMTIRR274 pKa = 11.84FALGQKK280 pKa = 8.73WPPLALAAFSGEE292 pKa = 3.91IVKK295 pKa = 9.67MKK297 pKa = 10.77SLMLQYY303 pKa = 10.52RR304 pKa = 11.84RR305 pKa = 11.84LGDD308 pKa = 3.3RR309 pKa = 11.84AKK311 pKa = 10.92FMALLEE317 pKa = 4.23MSEE320 pKa = 4.33MMDD323 pKa = 3.59FAPSNFPLLYY333 pKa = 9.8SYY335 pKa = 11.8AMGIGSVQDD344 pKa = 3.55PQMRR348 pKa = 11.84GYY350 pKa = 9.17TFGRR354 pKa = 11.84PYY356 pKa = 10.92LNAAFYY362 pKa = 10.48QLGVEE367 pKa = 4.61TANQQQGSVDD377 pKa = 3.17KK378 pKa = 11.53DD379 pKa = 3.21MAAEE383 pKa = 4.42LGLTEE388 pKa = 4.17ADD390 pKa = 3.59KK391 pKa = 11.18RR392 pKa = 11.84AMAATVTRR400 pKa = 11.84LTTGRR405 pKa = 11.84GGLQGNMGINAMARR419 pKa = 11.84RR420 pKa = 11.84GQQLPQPQDD429 pKa = 3.21NVVEE433 pKa = 4.09EE434 pKa = 4.65DD435 pKa = 3.58EE436 pKa = 5.03EE437 pKa = 4.3EE438 pKa = 4.54DD439 pKa = 3.84EE440 pKa = 5.51EE441 pKa = 5.06EE442 pKa = 4.91EE443 pKa = 4.28EE444 pKa = 4.24PQNQEE449 pKa = 4.0RR450 pKa = 11.84EE451 pKa = 4.03VDD453 pKa = 3.49PEE455 pKa = 4.05LEE457 pKa = 3.83RR458 pKa = 11.84RR459 pKa = 11.84VALRR463 pKa = 11.84IAQDD467 pKa = 3.2QEE469 pKa = 3.29RR470 pKa = 11.84WARR473 pKa = 11.84RR474 pKa = 11.84LAEE477 pKa = 3.99IEE479 pKa = 3.95AEE481 pKa = 4.16RR482 pKa = 11.84VARR485 pKa = 11.84QAASHH490 pKa = 7.01PPQHH494 pKa = 7.12DD495 pKa = 3.44ATRR498 pKa = 11.84DD499 pKa = 3.62QIPDD503 pKa = 3.34QQEE506 pKa = 3.79YY507 pKa = 10.96DD508 pKa = 3.74SNLIEE513 pKa = 4.2
MM1 pKa = 7.63ASLFRR6 pKa = 11.84ALEE9 pKa = 4.01QFTLEE14 pKa = 3.83QDD16 pKa = 2.79QGGRR20 pKa = 11.84GNVGDD25 pKa = 4.39QPPEE29 pKa = 3.9TLQATIKK36 pKa = 10.49VFVINTPDD44 pKa = 2.96PRR46 pKa = 11.84LRR48 pKa = 11.84YY49 pKa = 10.36RR50 pKa = 11.84MMCFCLRR57 pKa = 11.84LIVSNSARR65 pKa = 11.84TGQRR69 pKa = 11.84HH70 pKa = 5.2GALMTLLSLPTAVMQNHH87 pKa = 5.7FRR89 pKa = 11.84TAEE92 pKa = 3.95RR93 pKa = 11.84SPDD96 pKa = 3.6CQIEE100 pKa = 4.28RR101 pKa = 11.84IEE103 pKa = 4.03VDD105 pKa = 3.04GFEE108 pKa = 4.54PGTYY112 pKa = 9.68RR113 pKa = 11.84IRR115 pKa = 11.84TNARR119 pKa = 11.84TPLTHH124 pKa = 6.63GEE126 pKa = 4.27VVALEE131 pKa = 4.17EE132 pKa = 4.17MADD135 pKa = 4.56DD136 pKa = 4.66IPEE139 pKa = 4.17ALANHH144 pKa = 6.27TPFVDD149 pKa = 3.4AQTEE153 pKa = 4.25LEE155 pKa = 4.45EE156 pKa = 5.24CDD158 pKa = 3.62EE159 pKa = 4.19MEE161 pKa = 4.74KK162 pKa = 10.56FLEE165 pKa = 5.01AIYY168 pKa = 9.24STLIQVWIMVTKK180 pKa = 10.91SMTAFDD186 pKa = 4.14QPTGSDD192 pKa = 3.06EE193 pKa = 4.23RR194 pKa = 11.84RR195 pKa = 11.84VAKK198 pKa = 9.97YY199 pKa = 8.5QQQGRR204 pKa = 11.84LNPHH208 pKa = 6.3YY209 pKa = 10.19LLQGEE214 pKa = 4.44VRR216 pKa = 11.84RR217 pKa = 11.84RR218 pKa = 11.84IQLCIRR224 pKa = 11.84QSLPIRR230 pKa = 11.84QFLVNEE236 pKa = 4.35LQTASNQGPITGKK249 pKa = 10.1YY250 pKa = 7.77YY251 pKa = 11.55AMVADD256 pKa = 3.46IGKK259 pKa = 10.03YY260 pKa = 9.88IMNAGMGGFFMTIRR274 pKa = 11.84FALGQKK280 pKa = 8.73WPPLALAAFSGEE292 pKa = 3.91IVKK295 pKa = 9.67MKK297 pKa = 10.77SLMLQYY303 pKa = 10.52RR304 pKa = 11.84RR305 pKa = 11.84LGDD308 pKa = 3.3RR309 pKa = 11.84AKK311 pKa = 10.92FMALLEE317 pKa = 4.23MSEE320 pKa = 4.33MMDD323 pKa = 3.59FAPSNFPLLYY333 pKa = 9.8SYY335 pKa = 11.8AMGIGSVQDD344 pKa = 3.55PQMRR348 pKa = 11.84GYY350 pKa = 9.17TFGRR354 pKa = 11.84PYY356 pKa = 10.92LNAAFYY362 pKa = 10.48QLGVEE367 pKa = 4.61TANQQQGSVDD377 pKa = 3.17KK378 pKa = 11.53DD379 pKa = 3.21MAAEE383 pKa = 4.42LGLTEE388 pKa = 4.17ADD390 pKa = 3.59KK391 pKa = 11.18RR392 pKa = 11.84AMAATVTRR400 pKa = 11.84LTTGRR405 pKa = 11.84GGLQGNMGINAMARR419 pKa = 11.84RR420 pKa = 11.84GQQLPQPQDD429 pKa = 3.21NVVEE433 pKa = 4.09EE434 pKa = 4.65DD435 pKa = 3.58EE436 pKa = 5.03EE437 pKa = 4.3EE438 pKa = 4.54DD439 pKa = 3.84EE440 pKa = 5.51EE441 pKa = 5.06EE442 pKa = 4.91EE443 pKa = 4.28EE444 pKa = 4.24PQNQEE449 pKa = 4.0RR450 pKa = 11.84EE451 pKa = 4.03VDD453 pKa = 3.49PEE455 pKa = 4.05LEE457 pKa = 3.83RR458 pKa = 11.84RR459 pKa = 11.84VALRR463 pKa = 11.84IAQDD467 pKa = 3.2QEE469 pKa = 3.29RR470 pKa = 11.84WARR473 pKa = 11.84RR474 pKa = 11.84LAEE477 pKa = 3.99IEE479 pKa = 3.95AEE481 pKa = 4.16RR482 pKa = 11.84VARR485 pKa = 11.84QAASHH490 pKa = 7.01PPQHH494 pKa = 7.12DD495 pKa = 3.44ATRR498 pKa = 11.84DD499 pKa = 3.62QIPDD503 pKa = 3.34QQEE506 pKa = 3.79YY507 pKa = 10.96DD508 pKa = 3.74SNLIEE513 pKa = 4.2
Molecular weight: 58.16 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D8WJ40|D8WJ40_9MONO Fusion glycoprotein F0 OS=Tuhoko virus 3 OX=798074 GN=F PE=3 SV=1
MM1 pKa = 8.02AGRR4 pKa = 11.84QATIPVPINFEE15 pKa = 4.1SPKK18 pKa = 10.51NYY20 pKa = 10.39LNAFPIVQADD30 pKa = 3.58PSVSGEE36 pKa = 3.79AGKK39 pKa = 10.23LLKK42 pKa = 10.27QIRR45 pKa = 11.84FKK47 pKa = 11.3DD48 pKa = 3.34LTLRR52 pKa = 11.84GSTEE56 pKa = 3.84APISFVNSYY65 pKa = 11.02GFIKK69 pKa = 10.27PLRR72 pKa = 11.84TRR74 pKa = 11.84EE75 pKa = 3.92EE76 pKa = 4.52FFSEE80 pKa = 3.8MHH82 pKa = 6.62KK83 pKa = 10.54PSQAPCLTACCLPFGAGPAIEE104 pKa = 5.06HH105 pKa = 7.27PDD107 pKa = 4.28KK108 pKa = 11.44IMDD111 pKa = 4.58DD112 pKa = 3.59LDD114 pKa = 3.5KK115 pKa = 11.59VFIVVRR121 pKa = 11.84KK122 pKa = 8.48SASTIEE128 pKa = 3.99EE129 pKa = 4.4CVFDD133 pKa = 3.97IRR135 pKa = 11.84KK136 pKa = 9.34LPSSLSRR143 pKa = 11.84HH144 pKa = 4.48QLAGNRR150 pKa = 11.84VLCVASDD157 pKa = 3.84KK158 pKa = 11.02YY159 pKa = 11.05IKK161 pKa = 10.46APCKK165 pKa = 9.03LTSGMDD171 pKa = 3.18YY172 pKa = 10.04TYY174 pKa = 11.03NIVFLSITHH183 pKa = 6.78CPPSQKK189 pKa = 9.82FRR191 pKa = 11.84VPMPIQSLRR200 pKa = 11.84AKK202 pKa = 10.31VMRR205 pKa = 11.84SVHH208 pKa = 6.84LEE210 pKa = 3.32IMIKK214 pKa = 9.99VDD216 pKa = 3.63CDD218 pKa = 3.53KK219 pKa = 11.31NSPITKK225 pKa = 9.97NLIYY229 pKa = 10.67DD230 pKa = 4.19PSNDD234 pKa = 2.29IWMASIWFHH243 pKa = 6.49LCNFYY248 pKa = 10.75KK249 pKa = 10.53GSKK252 pKa = 8.91PFKK255 pKa = 10.3EE256 pKa = 4.09YY257 pKa = 10.65DD258 pKa = 3.59DD259 pKa = 3.38QHH261 pKa = 6.17FASKK265 pKa = 9.8CRR267 pKa = 11.84AMGLEE272 pKa = 4.02VGLVDD277 pKa = 3.92FWGPTFLVKK286 pKa = 10.56AHH288 pKa = 6.08GKK290 pKa = 9.03IPHH293 pKa = 6.69AARR296 pKa = 11.84PFFGKK301 pKa = 9.88HH302 pKa = 4.5GWVCHH307 pKa = 5.64PMMDD311 pKa = 4.2CAPAISKK318 pKa = 9.21SLWALSISILQVNAVLQASDD338 pKa = 4.18LNQMIRR344 pKa = 11.84MTDD347 pKa = 3.23VVFPKK352 pKa = 10.83VKK354 pKa = 10.17INPDD358 pKa = 2.73IAGVQKK364 pKa = 9.66TRR366 pKa = 11.84WNPVKK371 pKa = 10.67KK372 pKa = 10.35LVTIDD377 pKa = 3.08
MM1 pKa = 8.02AGRR4 pKa = 11.84QATIPVPINFEE15 pKa = 4.1SPKK18 pKa = 10.51NYY20 pKa = 10.39LNAFPIVQADD30 pKa = 3.58PSVSGEE36 pKa = 3.79AGKK39 pKa = 10.23LLKK42 pKa = 10.27QIRR45 pKa = 11.84FKK47 pKa = 11.3DD48 pKa = 3.34LTLRR52 pKa = 11.84GSTEE56 pKa = 3.84APISFVNSYY65 pKa = 11.02GFIKK69 pKa = 10.27PLRR72 pKa = 11.84TRR74 pKa = 11.84EE75 pKa = 3.92EE76 pKa = 4.52FFSEE80 pKa = 3.8MHH82 pKa = 6.62KK83 pKa = 10.54PSQAPCLTACCLPFGAGPAIEE104 pKa = 5.06HH105 pKa = 7.27PDD107 pKa = 4.28KK108 pKa = 11.44IMDD111 pKa = 4.58DD112 pKa = 3.59LDD114 pKa = 3.5KK115 pKa = 11.59VFIVVRR121 pKa = 11.84KK122 pKa = 8.48SASTIEE128 pKa = 3.99EE129 pKa = 4.4CVFDD133 pKa = 3.97IRR135 pKa = 11.84KK136 pKa = 9.34LPSSLSRR143 pKa = 11.84HH144 pKa = 4.48QLAGNRR150 pKa = 11.84VLCVASDD157 pKa = 3.84KK158 pKa = 11.02YY159 pKa = 11.05IKK161 pKa = 10.46APCKK165 pKa = 9.03LTSGMDD171 pKa = 3.18YY172 pKa = 10.04TYY174 pKa = 11.03NIVFLSITHH183 pKa = 6.78CPPSQKK189 pKa = 9.82FRR191 pKa = 11.84VPMPIQSLRR200 pKa = 11.84AKK202 pKa = 10.31VMRR205 pKa = 11.84SVHH208 pKa = 6.84LEE210 pKa = 3.32IMIKK214 pKa = 9.99VDD216 pKa = 3.63CDD218 pKa = 3.53KK219 pKa = 11.31NSPITKK225 pKa = 9.97NLIYY229 pKa = 10.67DD230 pKa = 4.19PSNDD234 pKa = 2.29IWMASIWFHH243 pKa = 6.49LCNFYY248 pKa = 10.75KK249 pKa = 10.53GSKK252 pKa = 8.91PFKK255 pKa = 10.3EE256 pKa = 4.09YY257 pKa = 10.65DD258 pKa = 3.59DD259 pKa = 3.38QHH261 pKa = 6.17FASKK265 pKa = 9.8CRR267 pKa = 11.84AMGLEE272 pKa = 4.02VGLVDD277 pKa = 3.92FWGPTFLVKK286 pKa = 10.56AHH288 pKa = 6.08GKK290 pKa = 9.03IPHH293 pKa = 6.69AARR296 pKa = 11.84PFFGKK301 pKa = 9.88HH302 pKa = 4.5GWVCHH307 pKa = 5.64PMMDD311 pKa = 4.2CAPAISKK318 pKa = 9.21SLWALSISILQVNAVLQASDD338 pKa = 4.18LNQMIRR344 pKa = 11.84MTDD347 pKa = 3.23VVFPKK352 pKa = 10.83VKK354 pKa = 10.17INPDD358 pKa = 2.73IAGVQKK364 pKa = 9.66TRR366 pKa = 11.84WNPVKK371 pKa = 10.67KK372 pKa = 10.35LVTIDD377 pKa = 3.08
Molecular weight: 42.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
4925 |
244 |
2271 |
703.6 |
78.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.558 ± 0.89 | 2.091 ± 0.359 |
4.731 ± 0.523 | 5.401 ± 0.588 |
3.492 ± 0.283 | 5.036 ± 0.366 |
2.213 ± 0.362 | 7.777 ± 0.638 |
4.832 ± 0.725 | 10.457 ± 1.349 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.944 ± 0.336 | 5.137 ± 0.472 |
5.442 ± 0.78 | 4.65 ± 0.849 |
4.995 ± 0.456 | 8.751 ± 0.694 |
6.112 ± 0.464 | 5.442 ± 0.448 |
1.015 ± 0.161 | 2.924 ± 0.406 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
