
Apis mellifera associated microvirus 30
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 7.46
Get precalculated fractions of proteins
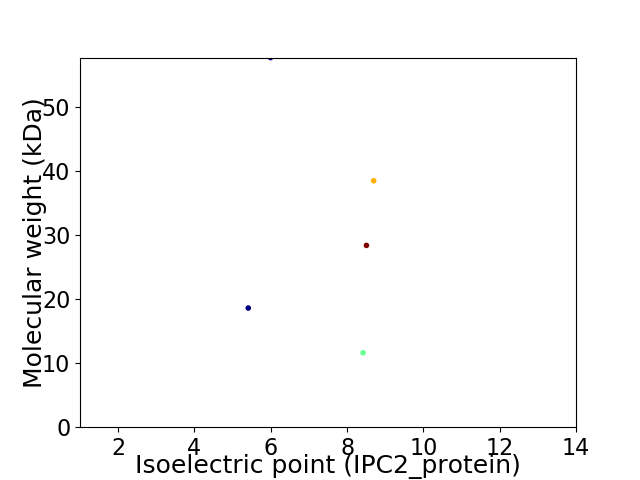
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3S8UTW0|A0A3S8UTW0_9VIRU Replication initiator protein OS=Apis mellifera associated microvirus 30 OX=2494759 PE=4 SV=1
MM1 pKa = 7.13KK2 pKa = 10.54HH3 pKa = 6.55EE4 pKa = 4.32ILLTNGEE11 pKa = 4.21IKK13 pKa = 10.76LPALQKK19 pKa = 10.21RR20 pKa = 11.84ARR22 pKa = 11.84HH23 pKa = 5.29YY24 pKa = 11.79ADD26 pKa = 5.03LDD28 pKa = 3.79YY29 pKa = 11.45TNTEE33 pKa = 4.15VQQHH37 pKa = 6.03FKK39 pKa = 11.16DD40 pKa = 3.38EE41 pKa = 4.18CDD43 pKa = 3.0INNIVKK49 pKa = 10.58RR50 pKa = 11.84LGPNGLAEE58 pKa = 5.37LIKK61 pKa = 10.04IDD63 pKa = 3.67PKK65 pKa = 10.87QYY67 pKa = 11.08GDD69 pKa = 3.61FSEE72 pKa = 4.68VPTYY76 pKa = 10.44QEE78 pKa = 3.6SLLIVSKK85 pKa = 10.92AQAQFDD91 pKa = 4.4ALPADD96 pKa = 3.52LRR98 pKa = 11.84DD99 pKa = 3.74RR100 pKa = 11.84FSNDD104 pKa = 2.75PAKK107 pKa = 10.5FLEE110 pKa = 4.49FTSNKK115 pKa = 9.55EE116 pKa = 3.98NLPEE120 pKa = 4.16MIKK123 pKa = 10.71LGLATEE129 pKa = 5.61RR130 pKa = 11.84IPNPPSDD137 pKa = 5.39LEE139 pKa = 4.01ILTNEE144 pKa = 4.47LKK146 pKa = 10.53RR147 pKa = 11.84RR148 pKa = 11.84NDD150 pKa = 2.99IDD152 pKa = 3.75SKK154 pKa = 11.34KK155 pKa = 9.83EE156 pKa = 4.0SKK158 pKa = 10.27PSSSTT163 pKa = 3.2
MM1 pKa = 7.13KK2 pKa = 10.54HH3 pKa = 6.55EE4 pKa = 4.32ILLTNGEE11 pKa = 4.21IKK13 pKa = 10.76LPALQKK19 pKa = 10.21RR20 pKa = 11.84ARR22 pKa = 11.84HH23 pKa = 5.29YY24 pKa = 11.79ADD26 pKa = 5.03LDD28 pKa = 3.79YY29 pKa = 11.45TNTEE33 pKa = 4.15VQQHH37 pKa = 6.03FKK39 pKa = 11.16DD40 pKa = 3.38EE41 pKa = 4.18CDD43 pKa = 3.0INNIVKK49 pKa = 10.58RR50 pKa = 11.84LGPNGLAEE58 pKa = 5.37LIKK61 pKa = 10.04IDD63 pKa = 3.67PKK65 pKa = 10.87QYY67 pKa = 11.08GDD69 pKa = 3.61FSEE72 pKa = 4.68VPTYY76 pKa = 10.44QEE78 pKa = 3.6SLLIVSKK85 pKa = 10.92AQAQFDD91 pKa = 4.4ALPADD96 pKa = 3.52LRR98 pKa = 11.84DD99 pKa = 3.74RR100 pKa = 11.84FSNDD104 pKa = 2.75PAKK107 pKa = 10.5FLEE110 pKa = 4.49FTSNKK115 pKa = 9.55EE116 pKa = 3.98NLPEE120 pKa = 4.16MIKK123 pKa = 10.71LGLATEE129 pKa = 5.61RR130 pKa = 11.84IPNPPSDD137 pKa = 5.39LEE139 pKa = 4.01ILTNEE144 pKa = 4.47LKK146 pKa = 10.53RR147 pKa = 11.84RR148 pKa = 11.84NDD150 pKa = 2.99IDD152 pKa = 3.75SKK154 pKa = 11.34KK155 pKa = 9.83EE156 pKa = 4.0SKK158 pKa = 10.27PSSSTT163 pKa = 3.2
Molecular weight: 18.58 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3S8UTW0|A0A3S8UTW0_9VIRU Replication initiator protein OS=Apis mellifera associated microvirus 30 OX=2494759 PE=4 SV=1
MM1 pKa = 7.46KK2 pKa = 9.94CLHH5 pKa = 6.34PVRR8 pKa = 11.84YY9 pKa = 9.21YY10 pKa = 10.81RR11 pKa = 11.84SRR13 pKa = 11.84DD14 pKa = 3.32SKK16 pKa = 11.48GNLTIHH22 pKa = 6.88HH23 pKa = 6.79SPNGVNVWSNGLRR36 pKa = 11.84PCGYY40 pKa = 9.53CAHH43 pKa = 6.66CRR45 pKa = 11.84LTKK48 pKa = 9.69AQSWAIRR55 pKa = 11.84MVHH58 pKa = 6.3HH59 pKa = 6.69AQMHH63 pKa = 5.07EE64 pKa = 3.91QSSFITLTYY73 pKa = 10.54DD74 pKa = 5.29DD75 pKa = 5.59KK76 pKa = 11.64NLPQNGSLNYY86 pKa = 9.69DD87 pKa = 3.32HH88 pKa = 7.0VVNFIKK94 pKa = 10.72RR95 pKa = 11.84LRR97 pKa = 11.84KK98 pKa = 9.4RR99 pKa = 11.84LHH101 pKa = 6.47SDD103 pKa = 2.84STNISFYY110 pKa = 10.82RR111 pKa = 11.84VGEE114 pKa = 4.1YY115 pKa = 10.43GGEE118 pKa = 4.01SHH120 pKa = 6.96RR121 pKa = 11.84PHH123 pKa = 5.92YY124 pKa = 10.5HH125 pKa = 7.22LILFGYY131 pKa = 10.27NFTEE135 pKa = 4.59RR136 pKa = 11.84IKK138 pKa = 11.26YY139 pKa = 10.08KK140 pKa = 10.5GVTNEE145 pKa = 4.8RR146 pKa = 11.84IVSSKK151 pKa = 10.82KK152 pKa = 10.15DD153 pKa = 3.01DD154 pKa = 3.27RR155 pKa = 11.84TYY157 pKa = 11.2YY158 pKa = 10.47KK159 pKa = 10.7SSFLTDD165 pKa = 3.04CWGHH169 pKa = 5.79GFADD173 pKa = 4.12LGDD176 pKa = 3.6VDD178 pKa = 6.56FATCQYY184 pKa = 7.17TAKK187 pKa = 10.68YY188 pKa = 9.99VMKK191 pKa = 10.56KK192 pKa = 10.18LYY194 pKa = 10.69APTAEE199 pKa = 3.96HH200 pKa = 5.94STWLEE205 pKa = 4.09KK206 pKa = 10.57EE207 pKa = 3.86RR208 pKa = 11.84SSCSKK213 pKa = 10.25RR214 pKa = 11.84IPIGTTWIQKK224 pKa = 8.34YY225 pKa = 7.3WQDD228 pKa = 3.99VYY230 pKa = 10.79PHH232 pKa = 7.64DD233 pKa = 4.34YY234 pKa = 10.71VLHH237 pKa = 6.95DD238 pKa = 3.94GKK240 pKa = 10.77KK241 pKa = 8.19YY242 pKa = 10.29KK243 pKa = 10.19PPRR246 pKa = 11.84FYY248 pKa = 11.27DD249 pKa = 2.94EE250 pKa = 4.29WLEE253 pKa = 4.05KK254 pKa = 10.47NQPLTYY260 pKa = 10.7AKK262 pKa = 10.5VKK264 pKa = 9.98LQRR267 pKa = 11.84EE268 pKa = 4.06EE269 pKa = 4.53SEE271 pKa = 4.15NDD273 pKa = 3.36VIDD276 pKa = 4.9IRR278 pKa = 11.84DD279 pKa = 3.78LHH281 pKa = 6.39AKK283 pKa = 9.93HH284 pKa = 6.52YY285 pKa = 10.68IRR287 pKa = 11.84IQKK290 pKa = 9.3QEE292 pKa = 3.56QFKK295 pKa = 10.89RR296 pKa = 11.84DD297 pKa = 4.4GIPHH301 pKa = 7.76DD302 pKa = 3.91LTLDD306 pKa = 3.95KK307 pKa = 10.89ILLEE311 pKa = 4.55RR312 pKa = 11.84KK313 pKa = 9.68LSTLHH318 pKa = 7.46DD319 pKa = 3.49IAKK322 pKa = 10.69GNLL325 pKa = 3.23
MM1 pKa = 7.46KK2 pKa = 9.94CLHH5 pKa = 6.34PVRR8 pKa = 11.84YY9 pKa = 9.21YY10 pKa = 10.81RR11 pKa = 11.84SRR13 pKa = 11.84DD14 pKa = 3.32SKK16 pKa = 11.48GNLTIHH22 pKa = 6.88HH23 pKa = 6.79SPNGVNVWSNGLRR36 pKa = 11.84PCGYY40 pKa = 9.53CAHH43 pKa = 6.66CRR45 pKa = 11.84LTKK48 pKa = 9.69AQSWAIRR55 pKa = 11.84MVHH58 pKa = 6.3HH59 pKa = 6.69AQMHH63 pKa = 5.07EE64 pKa = 3.91QSSFITLTYY73 pKa = 10.54DD74 pKa = 5.29DD75 pKa = 5.59KK76 pKa = 11.64NLPQNGSLNYY86 pKa = 9.69DD87 pKa = 3.32HH88 pKa = 7.0VVNFIKK94 pKa = 10.72RR95 pKa = 11.84LRR97 pKa = 11.84KK98 pKa = 9.4RR99 pKa = 11.84LHH101 pKa = 6.47SDD103 pKa = 2.84STNISFYY110 pKa = 10.82RR111 pKa = 11.84VGEE114 pKa = 4.1YY115 pKa = 10.43GGEE118 pKa = 4.01SHH120 pKa = 6.96RR121 pKa = 11.84PHH123 pKa = 5.92YY124 pKa = 10.5HH125 pKa = 7.22LILFGYY131 pKa = 10.27NFTEE135 pKa = 4.59RR136 pKa = 11.84IKK138 pKa = 11.26YY139 pKa = 10.08KK140 pKa = 10.5GVTNEE145 pKa = 4.8RR146 pKa = 11.84IVSSKK151 pKa = 10.82KK152 pKa = 10.15DD153 pKa = 3.01DD154 pKa = 3.27RR155 pKa = 11.84TYY157 pKa = 11.2YY158 pKa = 10.47KK159 pKa = 10.7SSFLTDD165 pKa = 3.04CWGHH169 pKa = 5.79GFADD173 pKa = 4.12LGDD176 pKa = 3.6VDD178 pKa = 6.56FATCQYY184 pKa = 7.17TAKK187 pKa = 10.68YY188 pKa = 9.99VMKK191 pKa = 10.56KK192 pKa = 10.18LYY194 pKa = 10.69APTAEE199 pKa = 3.96HH200 pKa = 5.94STWLEE205 pKa = 4.09KK206 pKa = 10.57EE207 pKa = 3.86RR208 pKa = 11.84SSCSKK213 pKa = 10.25RR214 pKa = 11.84IPIGTTWIQKK224 pKa = 8.34YY225 pKa = 7.3WQDD228 pKa = 3.99VYY230 pKa = 10.79PHH232 pKa = 7.64DD233 pKa = 4.34YY234 pKa = 10.71VLHH237 pKa = 6.95DD238 pKa = 3.94GKK240 pKa = 10.77KK241 pKa = 8.19YY242 pKa = 10.29KK243 pKa = 10.19PPRR246 pKa = 11.84FYY248 pKa = 11.27DD249 pKa = 2.94EE250 pKa = 4.29WLEE253 pKa = 4.05KK254 pKa = 10.47NQPLTYY260 pKa = 10.7AKK262 pKa = 10.5VKK264 pKa = 9.98LQRR267 pKa = 11.84EE268 pKa = 4.06EE269 pKa = 4.53SEE271 pKa = 4.15NDD273 pKa = 3.36VIDD276 pKa = 4.9IRR278 pKa = 11.84DD279 pKa = 3.78LHH281 pKa = 6.39AKK283 pKa = 9.93HH284 pKa = 6.52YY285 pKa = 10.68IRR287 pKa = 11.84IQKK290 pKa = 9.3QEE292 pKa = 3.56QFKK295 pKa = 10.89RR296 pKa = 11.84DD297 pKa = 4.4GIPHH301 pKa = 7.76DD302 pKa = 3.91LTLDD306 pKa = 3.95KK307 pKa = 10.89ILLEE311 pKa = 4.55RR312 pKa = 11.84KK313 pKa = 9.68LSTLHH318 pKa = 7.46DD319 pKa = 3.49IAKK322 pKa = 10.69GNLL325 pKa = 3.23
Molecular weight: 38.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1377 |
101 |
522 |
275.4 |
30.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.424 ± 2.232 | 0.944 ± 0.358 |
5.882 ± 0.483 | 4.938 ± 0.705 |
3.631 ± 0.527 | 6.245 ± 0.794 |
2.832 ± 0.905 | 5.374 ± 0.395 |
6.245 ± 1.501 | 7.553 ± 0.772 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.033 ± 0.305 | 5.737 ± 0.557 |
5.374 ± 1.003 | 4.575 ± 0.494 |
5.664 ± 0.341 | 7.262 ± 0.31 |
6.609 ± 0.655 | 5.011 ± 1.022 |
1.38 ± 0.268 | 4.285 ± 0.689 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
