
Streptococcus satellite phage Javan307
Taxonomy: Viruses; unclassified bacterial viruses
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
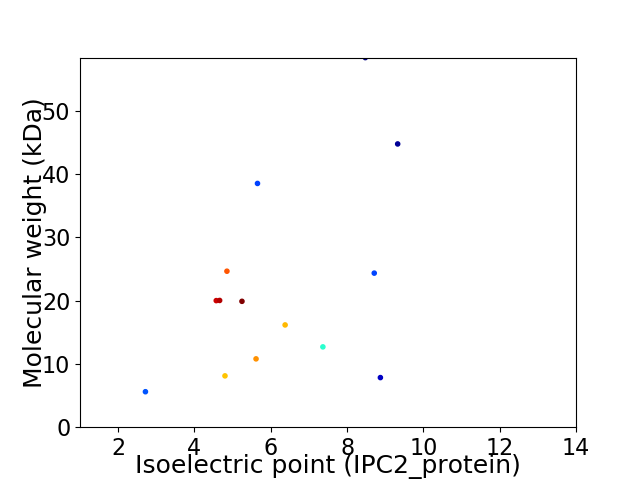
Virtual 2D-PAGE plot for 14 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
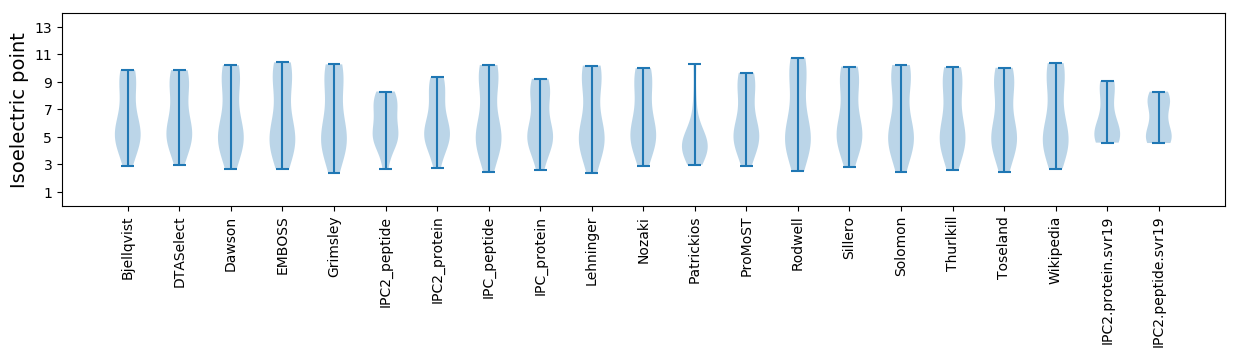
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4D5ZIV5|A0A4D5ZIV5_9VIRU Uncharacterized protein OS=Streptococcus satellite phage Javan307 OX=2558626 GN=JavanS307_0007 PE=4 SV=1
MM1 pKa = 7.54IADD4 pKa = 3.78KK5 pKa = 11.39LEE7 pKa = 4.85AISTNLEE14 pKa = 4.11EE15 pKa = 4.13IQEE18 pKa = 4.13ILVGNGTALAKK29 pKa = 10.45RR30 pKa = 11.84EE31 pKa = 3.86ISKK34 pKa = 10.38VLNDD38 pKa = 4.47LDD40 pKa = 5.59DD41 pKa = 4.4IYY43 pKa = 11.79NEE45 pKa = 4.13LTSTEE50 pKa = 3.99YY51 pKa = 10.82HH52 pKa = 5.9EE53 pKa = 4.26QQEE56 pKa = 4.54TLKK59 pKa = 10.72EE60 pKa = 3.7QTEE63 pKa = 4.21RR64 pKa = 11.84MKK66 pKa = 10.83QRR68 pKa = 11.84VVSEE72 pKa = 4.31VIGEE76 pKa = 4.27LEE78 pKa = 3.68WRR80 pKa = 11.84EE81 pKa = 3.45YY82 pKa = 11.05SYY84 pKa = 11.97YY85 pKa = 10.94KK86 pKa = 10.66NFWGDD91 pKa = 3.24MDD93 pKa = 5.26LLKK96 pKa = 10.88CLSNFDD102 pKa = 3.56GFLFISTYY110 pKa = 10.01LAEE113 pKa = 4.1ITKK116 pKa = 10.6EE117 pKa = 3.51NDD119 pKa = 2.93YY120 pKa = 11.52PMDD123 pKa = 4.03KK124 pKa = 10.74NQVLNYY130 pKa = 9.47VWEE133 pKa = 4.22LLAIDD138 pKa = 3.6IAKK141 pKa = 10.3KK142 pKa = 10.58KK143 pKa = 9.91RR144 pKa = 11.84DD145 pKa = 3.7KK146 pKa = 11.36KK147 pKa = 10.98NLLDD151 pKa = 3.74LWTSSIEE158 pKa = 4.03YY159 pKa = 8.46TRR161 pKa = 11.84GMMIYY166 pKa = 10.2EE167 pKa = 4.29EE168 pKa = 4.23NN169 pKa = 3.33
MM1 pKa = 7.54IADD4 pKa = 3.78KK5 pKa = 11.39LEE7 pKa = 4.85AISTNLEE14 pKa = 4.11EE15 pKa = 4.13IQEE18 pKa = 4.13ILVGNGTALAKK29 pKa = 10.45RR30 pKa = 11.84EE31 pKa = 3.86ISKK34 pKa = 10.38VLNDD38 pKa = 4.47LDD40 pKa = 5.59DD41 pKa = 4.4IYY43 pKa = 11.79NEE45 pKa = 4.13LTSTEE50 pKa = 3.99YY51 pKa = 10.82HH52 pKa = 5.9EE53 pKa = 4.26QQEE56 pKa = 4.54TLKK59 pKa = 10.72EE60 pKa = 3.7QTEE63 pKa = 4.21RR64 pKa = 11.84MKK66 pKa = 10.83QRR68 pKa = 11.84VVSEE72 pKa = 4.31VIGEE76 pKa = 4.27LEE78 pKa = 3.68WRR80 pKa = 11.84EE81 pKa = 3.45YY82 pKa = 11.05SYY84 pKa = 11.97YY85 pKa = 10.94KK86 pKa = 10.66NFWGDD91 pKa = 3.24MDD93 pKa = 5.26LLKK96 pKa = 10.88CLSNFDD102 pKa = 3.56GFLFISTYY110 pKa = 10.01LAEE113 pKa = 4.1ITKK116 pKa = 10.6EE117 pKa = 3.51NDD119 pKa = 2.93YY120 pKa = 11.52PMDD123 pKa = 4.03KK124 pKa = 10.74NQVLNYY130 pKa = 9.47VWEE133 pKa = 4.22LLAIDD138 pKa = 3.6IAKK141 pKa = 10.3KK142 pKa = 10.58KK143 pKa = 9.91RR144 pKa = 11.84DD145 pKa = 3.7KK146 pKa = 11.36KK147 pKa = 10.98NLLDD151 pKa = 3.74LWTSSIEE158 pKa = 4.03YY159 pKa = 8.46TRR161 pKa = 11.84GMMIYY166 pKa = 10.2EE167 pKa = 4.29EE168 pKa = 4.23NN169 pKa = 3.33
Molecular weight: 20.02 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4D5ZJP7|A0A4D5ZJP7_9VIRU Uncharacterized protein OS=Streptococcus satellite phage Javan307 OX=2558626 GN=JavanS307_0003 PE=4 SV=1
MM1 pKa = 7.84KK2 pKa = 9.12ITEE5 pKa = 4.39VIKK8 pKa = 10.77KK9 pKa = 10.02DD10 pKa = 3.19GSKK13 pKa = 10.33VYY15 pKa = 10.14RR16 pKa = 11.84ANVYY20 pKa = 10.6LGVDD24 pKa = 3.13QVTGKK29 pKa = 10.2KK30 pKa = 10.32VKK32 pKa = 9.81TKK34 pKa = 9.67VTGRR38 pKa = 11.84TQKK41 pKa = 10.07EE42 pKa = 4.39VKK44 pKa = 10.08QKK46 pKa = 11.27ANQEE50 pKa = 3.99KK51 pKa = 10.29IAFQKK56 pKa = 10.81AGSTRR61 pKa = 11.84QKK63 pKa = 11.02AITIKK68 pKa = 10.49NYY70 pKa = 10.3QEE72 pKa = 5.26LATLWWEE79 pKa = 4.3SYY81 pKa = 11.07KK82 pKa = 10.18NTVKK86 pKa = 10.71PNTQGNVRR94 pKa = 11.84ALIDD98 pKa = 3.5NHH100 pKa = 6.36ILPTFGEE107 pKa = 4.29YY108 pKa = 10.7KK109 pKa = 10.12LDD111 pKa = 3.72KK112 pKa = 10.14LTTPLIQSIINKK124 pKa = 9.52LADD127 pKa = 3.2KK128 pKa = 10.1TNRR131 pKa = 11.84GEE133 pKa = 4.1KK134 pKa = 10.44GAFLHH139 pKa = 6.19YY140 pKa = 10.82DD141 pKa = 4.02KK142 pKa = 11.05IHH144 pKa = 6.59ALNKK148 pKa = 10.18RR149 pKa = 11.84ILQYY153 pKa = 10.89GVTMQAIPSNPARR166 pKa = 11.84DD167 pKa = 3.48VVLPRR172 pKa = 11.84NTQKK176 pKa = 11.19AKK178 pKa = 8.99RR179 pKa = 11.84QKK181 pKa = 10.16VKK183 pKa = 10.6HH184 pKa = 5.86FEE186 pKa = 3.95NQEE189 pKa = 3.86LKK191 pKa = 10.93KK192 pKa = 10.77FLGYY196 pKa = 10.55LDD198 pKa = 5.12NLDD201 pKa = 3.56SDD203 pKa = 4.32RR204 pKa = 11.84YY205 pKa = 8.96RR206 pKa = 11.84YY207 pKa = 10.17YY208 pKa = 11.31YY209 pKa = 8.28EE210 pKa = 3.62TTLYY214 pKa = 10.88KK215 pKa = 10.46FLLATGCRR223 pKa = 11.84INEE226 pKa = 4.0ALALSWSDD234 pKa = 2.88IDD236 pKa = 4.45LDD238 pKa = 3.81NAVVHH243 pKa = 5.54ITKK246 pKa = 9.23TLNRR250 pKa = 11.84DD251 pKa = 3.49LEE253 pKa = 4.53INSPKK258 pKa = 10.62SKK260 pKa = 10.68ASYY263 pKa = 10.37RR264 pKa = 11.84DD265 pKa = 2.95IDD267 pKa = 3.41IDD269 pKa = 3.65QATVSMLKK277 pKa = 9.91QYY279 pKa = 11.27KK280 pKa = 9.69LRR282 pKa = 11.84QTKK285 pKa = 7.97EE286 pKa = 3.19AWKK289 pKa = 9.48IGQRR293 pKa = 11.84EE294 pKa = 4.21SVVFSDD300 pKa = 6.46FIHH303 pKa = 7.06EE304 pKa = 4.42YY305 pKa = 9.91PSSSRR310 pKa = 11.84LKK312 pKa = 10.58RR313 pKa = 11.84RR314 pKa = 11.84LQTHH318 pKa = 6.76FKK320 pKa = 10.46RR321 pKa = 11.84ADD323 pKa = 3.23VPNIGFHH330 pKa = 6.1GFRR333 pKa = 11.84HH334 pKa = 4.87THH336 pKa = 6.58ASLLLNSGIPYY347 pKa = 10.03KK348 pKa = 10.3EE349 pKa = 3.58LQYY352 pKa = 11.41RR353 pKa = 11.84LGHH356 pKa = 5.67STLSMTMDD364 pKa = 3.58IYY366 pKa = 11.68SHH368 pKa = 6.98LSKK371 pKa = 11.0EE372 pKa = 4.25NAKK375 pKa = 10.37KK376 pKa = 10.37AVSFYY381 pKa = 9.42EE382 pKa = 4.24TALKK386 pKa = 10.67ALL388 pKa = 4.15
MM1 pKa = 7.84KK2 pKa = 9.12ITEE5 pKa = 4.39VIKK8 pKa = 10.77KK9 pKa = 10.02DD10 pKa = 3.19GSKK13 pKa = 10.33VYY15 pKa = 10.14RR16 pKa = 11.84ANVYY20 pKa = 10.6LGVDD24 pKa = 3.13QVTGKK29 pKa = 10.2KK30 pKa = 10.32VKK32 pKa = 9.81TKK34 pKa = 9.67VTGRR38 pKa = 11.84TQKK41 pKa = 10.07EE42 pKa = 4.39VKK44 pKa = 10.08QKK46 pKa = 11.27ANQEE50 pKa = 3.99KK51 pKa = 10.29IAFQKK56 pKa = 10.81AGSTRR61 pKa = 11.84QKK63 pKa = 11.02AITIKK68 pKa = 10.49NYY70 pKa = 10.3QEE72 pKa = 5.26LATLWWEE79 pKa = 4.3SYY81 pKa = 11.07KK82 pKa = 10.18NTVKK86 pKa = 10.71PNTQGNVRR94 pKa = 11.84ALIDD98 pKa = 3.5NHH100 pKa = 6.36ILPTFGEE107 pKa = 4.29YY108 pKa = 10.7KK109 pKa = 10.12LDD111 pKa = 3.72KK112 pKa = 10.14LTTPLIQSIINKK124 pKa = 9.52LADD127 pKa = 3.2KK128 pKa = 10.1TNRR131 pKa = 11.84GEE133 pKa = 4.1KK134 pKa = 10.44GAFLHH139 pKa = 6.19YY140 pKa = 10.82DD141 pKa = 4.02KK142 pKa = 11.05IHH144 pKa = 6.59ALNKK148 pKa = 10.18RR149 pKa = 11.84ILQYY153 pKa = 10.89GVTMQAIPSNPARR166 pKa = 11.84DD167 pKa = 3.48VVLPRR172 pKa = 11.84NTQKK176 pKa = 11.19AKK178 pKa = 8.99RR179 pKa = 11.84QKK181 pKa = 10.16VKK183 pKa = 10.6HH184 pKa = 5.86FEE186 pKa = 3.95NQEE189 pKa = 3.86LKK191 pKa = 10.93KK192 pKa = 10.77FLGYY196 pKa = 10.55LDD198 pKa = 5.12NLDD201 pKa = 3.56SDD203 pKa = 4.32RR204 pKa = 11.84YY205 pKa = 8.96RR206 pKa = 11.84YY207 pKa = 10.17YY208 pKa = 11.31YY209 pKa = 8.28EE210 pKa = 3.62TTLYY214 pKa = 10.88KK215 pKa = 10.46FLLATGCRR223 pKa = 11.84INEE226 pKa = 4.0ALALSWSDD234 pKa = 2.88IDD236 pKa = 4.45LDD238 pKa = 3.81NAVVHH243 pKa = 5.54ITKK246 pKa = 9.23TLNRR250 pKa = 11.84DD251 pKa = 3.49LEE253 pKa = 4.53INSPKK258 pKa = 10.62SKK260 pKa = 10.68ASYY263 pKa = 10.37RR264 pKa = 11.84DD265 pKa = 2.95IDD267 pKa = 3.41IDD269 pKa = 3.65QATVSMLKK277 pKa = 9.91QYY279 pKa = 11.27KK280 pKa = 9.69LRR282 pKa = 11.84QTKK285 pKa = 7.97EE286 pKa = 3.19AWKK289 pKa = 9.48IGQRR293 pKa = 11.84EE294 pKa = 4.21SVVFSDD300 pKa = 6.46FIHH303 pKa = 7.06EE304 pKa = 4.42YY305 pKa = 9.91PSSSRR310 pKa = 11.84LKK312 pKa = 10.58RR313 pKa = 11.84RR314 pKa = 11.84LQTHH318 pKa = 6.76FKK320 pKa = 10.46RR321 pKa = 11.84ADD323 pKa = 3.23VPNIGFHH330 pKa = 6.1GFRR333 pKa = 11.84HH334 pKa = 4.87THH336 pKa = 6.58ASLLLNSGIPYY347 pKa = 10.03KK348 pKa = 10.3EE349 pKa = 3.58LQYY352 pKa = 11.41RR353 pKa = 11.84LGHH356 pKa = 5.67STLSMTMDD364 pKa = 3.58IYY366 pKa = 11.68SHH368 pKa = 6.98LSKK371 pKa = 11.0EE372 pKa = 4.25NAKK375 pKa = 10.37KK376 pKa = 10.37AVSFYY381 pKa = 9.42EE382 pKa = 4.24TALKK386 pKa = 10.67ALL388 pKa = 4.15
Molecular weight: 44.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2684 |
48 |
507 |
191.7 |
22.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.179 ± 0.381 | 0.261 ± 0.066 |
6.11 ± 0.498 | 8.793 ± 0.822 |
4.173 ± 0.381 | 4.881 ± 0.356 |
1.528 ± 0.279 | 7.004 ± 0.451 |
8.979 ± 0.502 | 10.656 ± 0.817 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.496 ± 0.339 | 5.365 ± 0.569 |
2.571 ± 0.583 | 4.322 ± 0.389 |
5.44 ± 0.357 | 5.775 ± 0.281 |
6.259 ± 0.468 | 4.471 ± 0.392 |
1.043 ± 0.156 | 4.694 ± 0.279 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
