
Canis familiaris papillomavirus 15
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Chipapillomavirus; Chipapillomavirus 3
Average proteome isoelectric point is 6.04
Get precalculated fractions of proteins
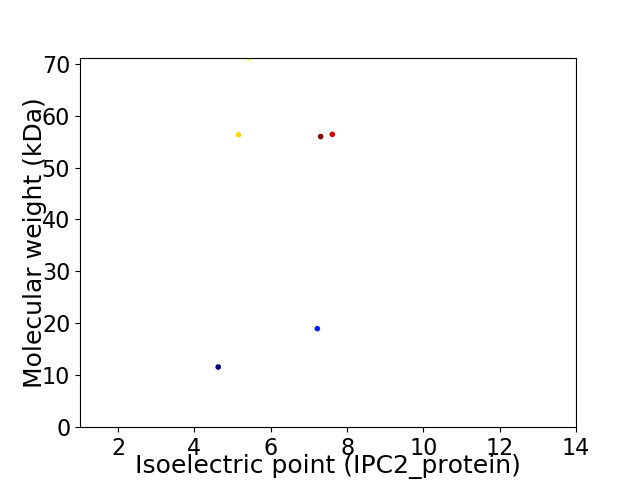
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
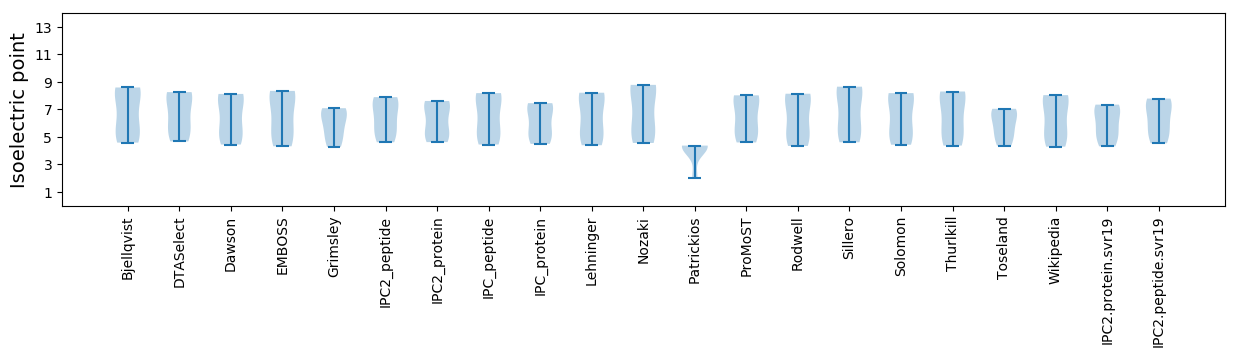
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|L0CKP3|L0CKP3_9PAPI Protein E7 OS=Canis familiaris papillomavirus 15 OX=1272519 GN=E7 PE=3 SV=1
MM1 pKa = 7.31IGKK4 pKa = 9.33GATLKK9 pKa = 10.95DD10 pKa = 3.3IVLEE14 pKa = 4.16EE15 pKa = 4.24QPCPYY20 pKa = 10.65DD21 pKa = 3.73EE22 pKa = 5.35LYY24 pKa = 10.79CDD26 pKa = 4.09EE27 pKa = 4.89EE28 pKa = 4.62LPPEE32 pKa = 4.39DD33 pKa = 4.17EE34 pKa = 4.72EE35 pKa = 4.24EE36 pKa = 4.66AEE38 pKa = 5.6QEE40 pKa = 4.4TQDD43 pKa = 3.21IAPYY47 pKa = 10.13RR48 pKa = 11.84IQTLCGHH55 pKa = 6.89CDD57 pKa = 2.69RR58 pKa = 11.84GIRR61 pKa = 11.84LVVLSTPHH69 pKa = 7.61GIRR72 pKa = 11.84TLEE75 pKa = 4.51DD76 pKa = 3.69LLLSCIGVCCPEE88 pKa = 4.2CASRR92 pKa = 11.84RR93 pKa = 11.84WRR95 pKa = 11.84FGRR98 pKa = 11.84AQHH101 pKa = 6.55GGG103 pKa = 3.34
MM1 pKa = 7.31IGKK4 pKa = 9.33GATLKK9 pKa = 10.95DD10 pKa = 3.3IVLEE14 pKa = 4.16EE15 pKa = 4.24QPCPYY20 pKa = 10.65DD21 pKa = 3.73EE22 pKa = 5.35LYY24 pKa = 10.79CDD26 pKa = 4.09EE27 pKa = 4.89EE28 pKa = 4.62LPPEE32 pKa = 4.39DD33 pKa = 4.17EE34 pKa = 4.72EE35 pKa = 4.24EE36 pKa = 4.66AEE38 pKa = 5.6QEE40 pKa = 4.4TQDD43 pKa = 3.21IAPYY47 pKa = 10.13RR48 pKa = 11.84IQTLCGHH55 pKa = 6.89CDD57 pKa = 2.69RR58 pKa = 11.84GIRR61 pKa = 11.84LVVLSTPHH69 pKa = 7.61GIRR72 pKa = 11.84TLEE75 pKa = 4.51DD76 pKa = 3.69LLLSCIGVCCPEE88 pKa = 4.2CASRR92 pKa = 11.84RR93 pKa = 11.84WRR95 pKa = 11.84FGRR98 pKa = 11.84AQHH101 pKa = 6.55GGG103 pKa = 3.34
Molecular weight: 11.59 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|L0CKP3|L0CKP3_9PAPI Protein E7 OS=Canis familiaris papillomavirus 15 OX=1272519 GN=E7 PE=3 SV=1
MM1 pKa = 7.01EE2 pKa = 4.74TLSKK6 pKa = 10.58RR7 pKa = 11.84LNALQDD13 pKa = 3.6ALLNLYY19 pKa = 10.05EE20 pKa = 4.26EE21 pKa = 5.25ASDD24 pKa = 3.56EE25 pKa = 4.14LQRR28 pKa = 11.84QVDD31 pKa = 2.97HH32 pKa = 6.56WKK34 pKa = 10.06ILRR37 pKa = 11.84QEE39 pKa = 4.03NVLLHH44 pKa = 5.48YY45 pKa = 10.24ARR47 pKa = 11.84KK48 pKa = 10.05SGVLRR53 pKa = 11.84VGMQPVPPLQVSAEE67 pKa = 4.17RR68 pKa = 11.84AKK70 pKa = 10.69HH71 pKa = 5.48AIEE74 pKa = 3.95MQIVLQSLCDD84 pKa = 3.87SQWASEE90 pKa = 4.02TWTLADD96 pKa = 3.66TSRR99 pKa = 11.84EE100 pKa = 3.94LWTAAPQRR108 pKa = 11.84CFKK111 pKa = 10.64KK112 pKa = 9.58GACIVEE118 pKa = 4.32VIYY121 pKa = 11.16DD122 pKa = 4.33GDD124 pKa = 4.35PDD126 pKa = 3.86NCMQYY131 pKa = 8.85TLWSRR136 pKa = 11.84VYY138 pKa = 10.14YY139 pKa = 10.35QDD141 pKa = 6.12GSDD144 pKa = 3.24TWQMSRR150 pKa = 11.84SQVDD154 pKa = 3.56CVGIWFWDD162 pKa = 4.06CEE164 pKa = 4.03QKK166 pKa = 10.54RR167 pKa = 11.84YY168 pKa = 8.81YY169 pKa = 11.12VKK171 pKa = 10.49FANDD175 pKa = 3.15AARR178 pKa = 11.84YY179 pKa = 6.19GTTGTWEE186 pKa = 3.85VVYY189 pKa = 10.89NGEE192 pKa = 4.67TVTSSDD198 pKa = 3.87PVTSTTPPTDD208 pKa = 3.37PCGEE212 pKa = 3.83APVYY216 pKa = 10.19HH217 pKa = 6.83RR218 pKa = 11.84RR219 pKa = 11.84LSPHH223 pKa = 6.34GPSAGDD229 pKa = 3.37ANPPIEE235 pKa = 4.88RR236 pKa = 11.84GGGDD240 pKa = 3.54RR241 pKa = 11.84TPASTPEE248 pKa = 3.78TATQVSSTYY257 pKa = 10.0TSPLPDD263 pKa = 3.41TTSPSPRR270 pKa = 11.84PLGEE274 pKa = 3.96VEE276 pKa = 3.83GDD278 pKa = 3.03RR279 pKa = 11.84GGGGGRR285 pKa = 11.84GATAEE290 pKa = 4.22PVGSSPLAAAAIAATPVSVPRR311 pKa = 11.84VPLTPRR317 pKa = 11.84KK318 pKa = 9.2RR319 pKa = 11.84PRR321 pKa = 11.84GRR323 pKa = 11.84GPSEE327 pKa = 3.88PPCRR331 pKa = 11.84RR332 pKa = 11.84QPPPSPGEE340 pKa = 3.93PEE342 pKa = 4.07PAVDD346 pKa = 3.78PGLVPVCGRR355 pKa = 11.84PRR357 pKa = 11.84TPPITPQGGQPSPLTVPVAEE377 pKa = 4.31PRR379 pKa = 11.84PVGSLTPPGGGLPPSPAPPAAEE401 pKa = 4.15AARR404 pKa = 11.84EE405 pKa = 3.96PARR408 pKa = 11.84RR409 pKa = 11.84YY410 pKa = 9.16PPAGARR416 pKa = 11.84RR417 pKa = 11.84LAVPGGHH424 pKa = 7.17PNRR427 pKa = 11.84PGDD430 pKa = 3.9LLRR433 pKa = 11.84EE434 pKa = 4.42AGHH437 pKa = 7.02LPQTAVVLCGPANALKK453 pKa = 10.3CLRR456 pKa = 11.84YY457 pKa = 9.59RR458 pKa = 11.84LQHH461 pKa = 5.29HH462 pKa = 7.16HH463 pKa = 5.65GQKK466 pKa = 10.46FLTVSTTWYY475 pKa = 8.59WAGQGSSRR483 pKa = 11.84LGPARR488 pKa = 11.84IMLTFRR494 pKa = 11.84DD495 pKa = 4.1SQQQTDD501 pKa = 3.64FFQSVRR507 pKa = 11.84LPPSISRR514 pKa = 11.84APSIFSVV521 pKa = 3.37
MM1 pKa = 7.01EE2 pKa = 4.74TLSKK6 pKa = 10.58RR7 pKa = 11.84LNALQDD13 pKa = 3.6ALLNLYY19 pKa = 10.05EE20 pKa = 4.26EE21 pKa = 5.25ASDD24 pKa = 3.56EE25 pKa = 4.14LQRR28 pKa = 11.84QVDD31 pKa = 2.97HH32 pKa = 6.56WKK34 pKa = 10.06ILRR37 pKa = 11.84QEE39 pKa = 4.03NVLLHH44 pKa = 5.48YY45 pKa = 10.24ARR47 pKa = 11.84KK48 pKa = 10.05SGVLRR53 pKa = 11.84VGMQPVPPLQVSAEE67 pKa = 4.17RR68 pKa = 11.84AKK70 pKa = 10.69HH71 pKa = 5.48AIEE74 pKa = 3.95MQIVLQSLCDD84 pKa = 3.87SQWASEE90 pKa = 4.02TWTLADD96 pKa = 3.66TSRR99 pKa = 11.84EE100 pKa = 3.94LWTAAPQRR108 pKa = 11.84CFKK111 pKa = 10.64KK112 pKa = 9.58GACIVEE118 pKa = 4.32VIYY121 pKa = 11.16DD122 pKa = 4.33GDD124 pKa = 4.35PDD126 pKa = 3.86NCMQYY131 pKa = 8.85TLWSRR136 pKa = 11.84VYY138 pKa = 10.14YY139 pKa = 10.35QDD141 pKa = 6.12GSDD144 pKa = 3.24TWQMSRR150 pKa = 11.84SQVDD154 pKa = 3.56CVGIWFWDD162 pKa = 4.06CEE164 pKa = 4.03QKK166 pKa = 10.54RR167 pKa = 11.84YY168 pKa = 8.81YY169 pKa = 11.12VKK171 pKa = 10.49FANDD175 pKa = 3.15AARR178 pKa = 11.84YY179 pKa = 6.19GTTGTWEE186 pKa = 3.85VVYY189 pKa = 10.89NGEE192 pKa = 4.67TVTSSDD198 pKa = 3.87PVTSTTPPTDD208 pKa = 3.37PCGEE212 pKa = 3.83APVYY216 pKa = 10.19HH217 pKa = 6.83RR218 pKa = 11.84RR219 pKa = 11.84LSPHH223 pKa = 6.34GPSAGDD229 pKa = 3.37ANPPIEE235 pKa = 4.88RR236 pKa = 11.84GGGDD240 pKa = 3.54RR241 pKa = 11.84TPASTPEE248 pKa = 3.78TATQVSSTYY257 pKa = 10.0TSPLPDD263 pKa = 3.41TTSPSPRR270 pKa = 11.84PLGEE274 pKa = 3.96VEE276 pKa = 3.83GDD278 pKa = 3.03RR279 pKa = 11.84GGGGGRR285 pKa = 11.84GATAEE290 pKa = 4.22PVGSSPLAAAAIAATPVSVPRR311 pKa = 11.84VPLTPRR317 pKa = 11.84KK318 pKa = 9.2RR319 pKa = 11.84PRR321 pKa = 11.84GRR323 pKa = 11.84GPSEE327 pKa = 3.88PPCRR331 pKa = 11.84RR332 pKa = 11.84QPPPSPGEE340 pKa = 3.93PEE342 pKa = 4.07PAVDD346 pKa = 3.78PGLVPVCGRR355 pKa = 11.84PRR357 pKa = 11.84TPPITPQGGQPSPLTVPVAEE377 pKa = 4.31PRR379 pKa = 11.84PVGSLTPPGGGLPPSPAPPAAEE401 pKa = 4.15AARR404 pKa = 11.84EE405 pKa = 3.96PARR408 pKa = 11.84RR409 pKa = 11.84YY410 pKa = 9.16PPAGARR416 pKa = 11.84RR417 pKa = 11.84LAVPGGHH424 pKa = 7.17PNRR427 pKa = 11.84PGDD430 pKa = 3.9LLRR433 pKa = 11.84EE434 pKa = 4.42AGHH437 pKa = 7.02LPQTAVVLCGPANALKK453 pKa = 10.3CLRR456 pKa = 11.84YY457 pKa = 9.59RR458 pKa = 11.84LQHH461 pKa = 5.29HH462 pKa = 7.16HH463 pKa = 5.65GQKK466 pKa = 10.46FLTVSTTWYY475 pKa = 8.59WAGQGSSRR483 pKa = 11.84LGPARR488 pKa = 11.84IMLTFRR494 pKa = 11.84DD495 pKa = 4.1SQQQTDD501 pKa = 3.64FFQSVRR507 pKa = 11.84LPPSISRR514 pKa = 11.84APSIFSVV521 pKa = 3.37
Molecular weight: 56.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2439 |
103 |
626 |
406.5 |
45.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.888 ± 0.571 | 2.296 ± 0.707 |
6.355 ± 0.502 | 6.068 ± 0.557 |
3.772 ± 0.54 | 7.913 ± 0.908 |
2.091 ± 0.232 | 3.567 ± 0.44 |
4.018 ± 1.046 | 8.282 ± 0.651 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.845 ± 0.523 | 3.321 ± 0.906 |
7.954 ± 1.386 | 4.182 ± 0.668 |
7.298 ± 0.811 | 7.257 ± 0.531 |
5.74 ± 0.539 | 6.929 ± 0.692 |
1.517 ± 0.294 | 2.706 ± 0.241 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
