
Dehalogenimonas lykanthroporepellens (strain ATCC BAA-1523 / JCM 15061 / BL-DC-9)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Chloroflexi; Dehalococcoidia; Dehalogenimonas; Dehalogenimonas lykanthroporepellens
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
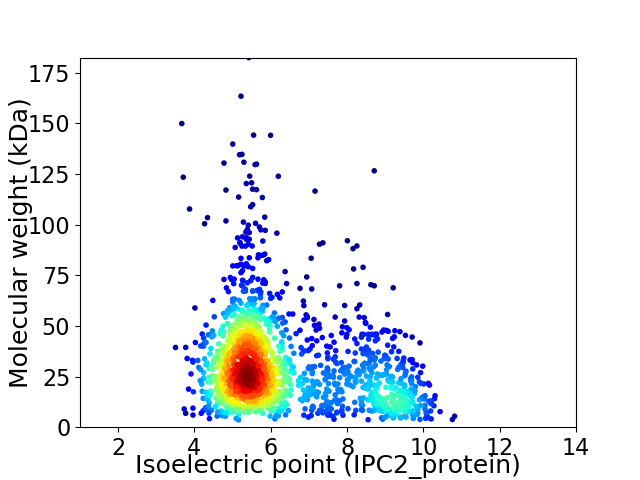
Virtual 2D-PAGE plot for 1619 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D8K447|D8K447_DEHLB Amidophosphoribosyltransferase OS=Dehalogenimonas lykanthroporepellens (strain ATCC BAA-1523 / JCM 15061 / BL-DC-9) OX=552811 GN=purF PE=3 SV=1
MM1 pKa = 7.49KK2 pKa = 10.32KK3 pKa = 9.73HH4 pKa = 5.91LRR6 pKa = 11.84KK7 pKa = 10.22LMMVGFVMVLAMSALVVGCDD27 pKa = 3.31STTDD31 pKa = 3.68EE32 pKa = 5.01PDD34 pKa = 3.08PTQSVNQKK42 pKa = 8.82PGQGVTMSPAKK53 pKa = 8.82PTWSTGWFQTEE64 pKa = 4.06VFIKK68 pKa = 10.5ALEE71 pKa = 3.87EE72 pKa = 3.62LGYY75 pKa = 10.72NVEE78 pKa = 5.0DD79 pKa = 6.08AIALDD84 pKa = 4.05NPIFYY89 pKa = 10.85NSVANGDD96 pKa = 3.27VDD98 pKa = 4.59FWADD102 pKa = 2.8GWFPLHH108 pKa = 6.1NQYY111 pKa = 11.24LPIIEE116 pKa = 4.59GKK118 pKa = 10.55AEE120 pKa = 4.19VVGEE124 pKa = 3.95FAMGGALQGYY134 pKa = 9.55LIDD137 pKa = 3.99KK138 pKa = 8.32ATAVEE143 pKa = 4.09YY144 pKa = 10.49GITSIEE150 pKa = 4.18DD151 pKa = 3.46FKK153 pKa = 11.56DD154 pKa = 3.36PEE156 pKa = 3.8IAALFDD162 pKa = 3.62TDD164 pKa = 4.6GDD166 pKa = 3.96GKK168 pKa = 10.96ANMVACPPGWGCEE181 pKa = 4.01TVINHH186 pKa = 6.0HH187 pKa = 5.89MEE189 pKa = 4.38AYY191 pKa = 9.83EE192 pKa = 4.06LADD195 pKa = 3.46WVNADD200 pKa = 3.28QAGYY204 pKa = 9.82DD205 pKa = 3.36VAMADD210 pKa = 3.33VVARR214 pKa = 11.84YY215 pKa = 8.59NAGEE219 pKa = 3.95PVFFYY224 pKa = 9.96TWTPNWTVNALVPGEE239 pKa = 4.07DD240 pKa = 4.01VVWLEE245 pKa = 4.49VPFSSLPADD254 pKa = 3.58QADD257 pKa = 4.21LEE259 pKa = 4.91DD260 pKa = 3.72EE261 pKa = 4.48TFVANLVGKK270 pKa = 10.3AGDD273 pKa = 3.82SEE275 pKa = 4.83PYY277 pKa = 10.48NMGWPANDD285 pKa = 3.04IQVVANSAFLDD296 pKa = 3.94ANPAAAYY303 pKa = 10.12LFEE306 pKa = 5.58NIQIPLEE313 pKa = 4.81DD314 pKa = 3.65IFEE317 pKa = 4.43QNYY320 pKa = 9.63QMYY323 pKa = 9.96QGEE326 pKa = 4.85DD327 pKa = 3.24RR328 pKa = 11.84PEE330 pKa = 5.9DD331 pKa = 3.38IVAHH335 pKa = 5.81AEE337 pKa = 3.41AWIAANQSTFDD348 pKa = 3.25AWIEE352 pKa = 4.05GAIDD356 pKa = 3.55AADD359 pKa = 3.07
MM1 pKa = 7.49KK2 pKa = 10.32KK3 pKa = 9.73HH4 pKa = 5.91LRR6 pKa = 11.84KK7 pKa = 10.22LMMVGFVMVLAMSALVVGCDD27 pKa = 3.31STTDD31 pKa = 3.68EE32 pKa = 5.01PDD34 pKa = 3.08PTQSVNQKK42 pKa = 8.82PGQGVTMSPAKK53 pKa = 8.82PTWSTGWFQTEE64 pKa = 4.06VFIKK68 pKa = 10.5ALEE71 pKa = 3.87EE72 pKa = 3.62LGYY75 pKa = 10.72NVEE78 pKa = 5.0DD79 pKa = 6.08AIALDD84 pKa = 4.05NPIFYY89 pKa = 10.85NSVANGDD96 pKa = 3.27VDD98 pKa = 4.59FWADD102 pKa = 2.8GWFPLHH108 pKa = 6.1NQYY111 pKa = 11.24LPIIEE116 pKa = 4.59GKK118 pKa = 10.55AEE120 pKa = 4.19VVGEE124 pKa = 3.95FAMGGALQGYY134 pKa = 9.55LIDD137 pKa = 3.99KK138 pKa = 8.32ATAVEE143 pKa = 4.09YY144 pKa = 10.49GITSIEE150 pKa = 4.18DD151 pKa = 3.46FKK153 pKa = 11.56DD154 pKa = 3.36PEE156 pKa = 3.8IAALFDD162 pKa = 3.62TDD164 pKa = 4.6GDD166 pKa = 3.96GKK168 pKa = 10.96ANMVACPPGWGCEE181 pKa = 4.01TVINHH186 pKa = 6.0HH187 pKa = 5.89MEE189 pKa = 4.38AYY191 pKa = 9.83EE192 pKa = 4.06LADD195 pKa = 3.46WVNADD200 pKa = 3.28QAGYY204 pKa = 9.82DD205 pKa = 3.36VAMADD210 pKa = 3.33VVARR214 pKa = 11.84YY215 pKa = 8.59NAGEE219 pKa = 3.95PVFFYY224 pKa = 9.96TWTPNWTVNALVPGEE239 pKa = 4.07DD240 pKa = 4.01VVWLEE245 pKa = 4.49VPFSSLPADD254 pKa = 3.58QADD257 pKa = 4.21LEE259 pKa = 4.91DD260 pKa = 3.72EE261 pKa = 4.48TFVANLVGKK270 pKa = 10.3AGDD273 pKa = 3.82SEE275 pKa = 4.83PYY277 pKa = 10.48NMGWPANDD285 pKa = 3.04IQVVANSAFLDD296 pKa = 3.94ANPAAAYY303 pKa = 10.12LFEE306 pKa = 5.58NIQIPLEE313 pKa = 4.81DD314 pKa = 3.65IFEE317 pKa = 4.43QNYY320 pKa = 9.63QMYY323 pKa = 9.96QGEE326 pKa = 4.85DD327 pKa = 3.24RR328 pKa = 11.84PEE330 pKa = 5.9DD331 pKa = 3.38IVAHH335 pKa = 5.81AEE337 pKa = 3.41AWIAANQSTFDD348 pKa = 3.25AWIEE352 pKa = 4.05GAIDD356 pKa = 3.55AADD359 pKa = 3.07
Molecular weight: 39.39 kDa
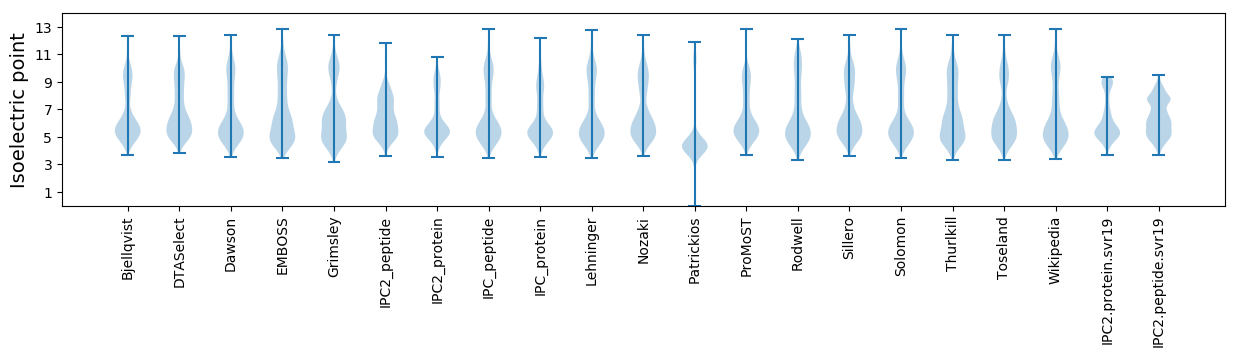
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D8K001|D8K001_DEHLB Lysine--tRNA ligase OS=Dehalogenimonas lykanthroporepellens (strain ATCC BAA-1523 / JCM 15061 / BL-DC-9) OX=552811 GN=lysS PE=3 SV=1
MM1 pKa = 7.42NLTLEE6 pKa = 4.24YY7 pKa = 10.32LLSVFIAALAVIQLGAVRR25 pKa = 11.84AGLRR29 pKa = 11.84GLYY32 pKa = 10.21LGGSRR37 pKa = 11.84LVACLVAVPCLLISGILFFTWNDD60 pKa = 3.39RR61 pKa = 11.84NPTGVIEE68 pKa = 4.6GAQQAGLFSLAVAAAAAFTVILGLITNRR96 pKa = 11.84RR97 pKa = 11.84MPADD101 pKa = 3.93RR102 pKa = 11.84PTSTEE107 pKa = 3.58GLEE110 pKa = 3.96ALKK113 pKa = 9.73RR114 pKa = 11.84TTFIHH119 pKa = 7.54AIRR122 pKa = 11.84SRR124 pKa = 11.84FWKK127 pKa = 10.5SS128 pKa = 2.65
MM1 pKa = 7.42NLTLEE6 pKa = 4.24YY7 pKa = 10.32LLSVFIAALAVIQLGAVRR25 pKa = 11.84AGLRR29 pKa = 11.84GLYY32 pKa = 10.21LGGSRR37 pKa = 11.84LVACLVAVPCLLISGILFFTWNDD60 pKa = 3.39RR61 pKa = 11.84NPTGVIEE68 pKa = 4.6GAQQAGLFSLAVAAAAAFTVILGLITNRR96 pKa = 11.84RR97 pKa = 11.84MPADD101 pKa = 3.93RR102 pKa = 11.84PTSTEE107 pKa = 3.58GLEE110 pKa = 3.96ALKK113 pKa = 9.73RR114 pKa = 11.84TTFIHH119 pKa = 7.54AIRR122 pKa = 11.84SRR124 pKa = 11.84FWKK127 pKa = 10.5SS128 pKa = 2.65
Molecular weight: 13.75 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
477317 |
31 |
1597 |
294.8 |
32.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.652 ± 0.083 | 1.103 ± 0.027 |
5.545 ± 0.046 | 6.754 ± 0.063 |
3.695 ± 0.043 | 8.105 ± 0.065 |
1.911 ± 0.024 | 6.405 ± 0.057 |
4.419 ± 0.057 | 9.948 ± 0.063 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.407 ± 0.031 | 3.328 ± 0.039 |
4.611 ± 0.034 | 3.145 ± 0.033 |
6.528 ± 0.066 | 5.684 ± 0.043 |
5.559 ± 0.049 | 7.339 ± 0.054 |
1.102 ± 0.026 | 2.76 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
