
Trypanosoma cruzi marinkellei
Taxonomy: cellular organisms; Eukaryota; Discoba; Euglenozoa; Kinetoplastea; Metakinetoplastina; Trypanosomatida; Trypanosomatidae; Trypanosoma; Schizotrypanum; Trypanosoma cruzi
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
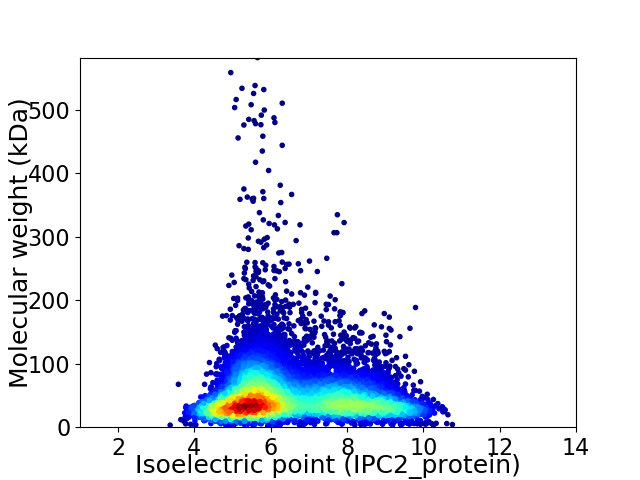
Virtual 2D-PAGE plot for 10052 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
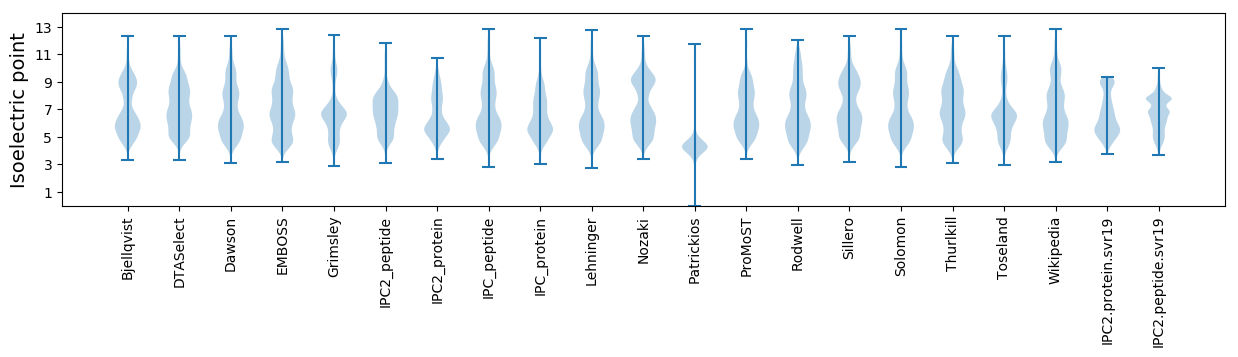
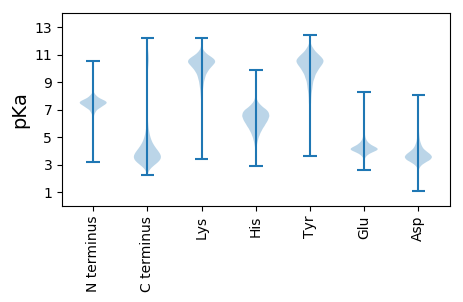
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K2NH72|K2NH72_TRYCR Dispersed gene family protein 1 (DGF-1) putative (Fragment) OS=Trypanosoma cruzi marinkellei OX=85056 GN=MOQ_007938 PE=4 SV=1
MM1 pKa = 6.85MTTCRR6 pKa = 11.84LLCALLVLALCCCPSVCGTEE26 pKa = 3.8VDD28 pKa = 3.92VNKK31 pKa = 10.94DD32 pKa = 3.29KK33 pKa = 11.6GNSSTPSQPGGTGVLGPGEE52 pKa = 4.28PQSLSQEE59 pKa = 4.08PGIQSGQSGEE69 pKa = 5.54DD70 pKa = 3.3GMQHH74 pKa = 5.33NTEE77 pKa = 4.17QVPGQPADD85 pKa = 3.33TALTEE90 pKa = 4.18NPNAGTPGPITEE102 pKa = 4.22QGPRR106 pKa = 11.84DD107 pKa = 3.84TDD109 pKa = 3.51TQEE112 pKa = 4.1AQSGDD117 pKa = 3.53TAASGQTDD125 pKa = 3.79TNVDD129 pKa = 3.64DD130 pKa = 4.3SAEE133 pKa = 4.27TTTANAPTTATTTTTAPEE151 pKa = 4.33APSTTTTEE159 pKa = 4.19APTTTTTRR167 pKa = 11.84APSRR171 pKa = 11.84LPEE174 pKa = 5.1IDD176 pKa = 3.88GSLSSSAWVIAPLLLAVSALAYY198 pKa = 6.39TTLGG202 pKa = 3.38
MM1 pKa = 6.85MTTCRR6 pKa = 11.84LLCALLVLALCCCPSVCGTEE26 pKa = 3.8VDD28 pKa = 3.92VNKK31 pKa = 10.94DD32 pKa = 3.29KK33 pKa = 11.6GNSSTPSQPGGTGVLGPGEE52 pKa = 4.28PQSLSQEE59 pKa = 4.08PGIQSGQSGEE69 pKa = 5.54DD70 pKa = 3.3GMQHH74 pKa = 5.33NTEE77 pKa = 4.17QVPGQPADD85 pKa = 3.33TALTEE90 pKa = 4.18NPNAGTPGPITEE102 pKa = 4.22QGPRR106 pKa = 11.84DD107 pKa = 3.84TDD109 pKa = 3.51TQEE112 pKa = 4.1AQSGDD117 pKa = 3.53TAASGQTDD125 pKa = 3.79TNVDD129 pKa = 3.64DD130 pKa = 4.3SAEE133 pKa = 4.27TTTANAPTTATTTTTAPEE151 pKa = 4.33APSTTTTEE159 pKa = 4.19APTTTTTRR167 pKa = 11.84APSRR171 pKa = 11.84LPEE174 pKa = 5.1IDD176 pKa = 3.88GSLSSSAWVIAPLLLAVSALAYY198 pKa = 6.39TTLGG202 pKa = 3.38
Molecular weight: 20.33 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K2NUY0|K2NUY0_TRYCR DNA-directed RNA polymerase subunit OS=Trypanosoma cruzi marinkellei OX=85056 GN=MOQ_003349 PE=3 SV=1
MM1 pKa = 8.11DD2 pKa = 3.94VAQFLNVRR10 pKa = 11.84VEE12 pKa = 3.93EE13 pKa = 4.08LAAFSALLFRR23 pKa = 11.84KK24 pKa = 10.11RR25 pKa = 11.84PRR27 pKa = 11.84HH28 pKa = 4.86EE29 pKa = 3.92NEE31 pKa = 4.04GAFSCTARR39 pKa = 11.84SVVHH43 pKa = 5.44YY44 pKa = 9.69RR45 pKa = 11.84RR46 pKa = 11.84PSSANVPLSDD56 pKa = 3.94VMRR59 pKa = 11.84LAKK62 pKa = 10.1FRR64 pKa = 11.84SRR66 pKa = 11.84RR67 pKa = 11.84SKK69 pKa = 10.66RR70 pKa = 11.84FVRR73 pKa = 11.84LLAAEE78 pKa = 4.58EE79 pKa = 4.48PPPATPKK86 pKa = 10.57AVEE89 pKa = 4.08EE90 pKa = 4.32EE91 pKa = 4.0KK92 pKa = 11.32GRR94 pKa = 11.84MQKK97 pKa = 9.91KK98 pKa = 9.55RR99 pKa = 11.84RR100 pKa = 11.84LLTYY104 pKa = 10.15RR105 pKa = 11.84AWCLLLRR112 pKa = 11.84RR113 pKa = 11.84RR114 pKa = 11.84RR115 pKa = 11.84RR116 pKa = 11.84FLLRR120 pKa = 11.84RR121 pKa = 11.84RR122 pKa = 11.84ATTRR126 pKa = 11.84RR127 pKa = 11.84RR128 pKa = 11.84IRR130 pKa = 11.84RR131 pKa = 11.84RR132 pKa = 11.84IISLRR137 pKa = 11.84YY138 pKa = 9.86GKK140 pKa = 7.89TQLPFCNCGSAAGAPPPSFPPCLSTQSSSSGGGRR174 pKa = 11.84TAVLWLPSHH183 pKa = 6.17WRR185 pKa = 11.84FCRR188 pKa = 11.84RR189 pKa = 11.84FHH191 pKa = 6.73YY192 pKa = 10.69SVFKK196 pKa = 10.55TRR198 pKa = 11.84INGCSSSPDD207 pKa = 3.51DD208 pKa = 4.56DD209 pKa = 4.41FSRR212 pKa = 11.84LVNVRR217 pKa = 11.84IAVPTKK223 pKa = 8.66SQRR226 pKa = 11.84KK227 pKa = 4.49QHH229 pKa = 6.18RR230 pKa = 11.84VLQRR234 pKa = 11.84WAARR238 pKa = 11.84LPSCCALGKK247 pKa = 9.85PPGRR251 pKa = 11.84IKK253 pKa = 10.52SKK255 pKa = 10.33PMAVPPPLCFAAEE268 pKa = 4.0RR269 pKa = 11.84SHH271 pKa = 5.42MCVWAIQPRR280 pKa = 11.84EE281 pKa = 4.0NGHH284 pKa = 7.09LLTADD289 pKa = 3.41EE290 pKa = 4.61VMDD293 pKa = 3.3ILMIHH298 pKa = 6.09SCKK301 pKa = 10.4APPVALHH308 pKa = 5.93TRR310 pKa = 11.84DD311 pKa = 4.1LATMAQTSVGSVPHH325 pKa = 5.71EE326 pKa = 4.17KK327 pKa = 9.71CWRR330 pKa = 11.84RR331 pKa = 11.84TEE333 pKa = 3.98SAVFYY338 pKa = 10.44GYY340 pKa = 8.58MWSLGKK346 pKa = 10.58AMSKK350 pKa = 10.47SKK352 pKa = 9.94WLLRR356 pKa = 11.84GDD358 pKa = 4.25TEE360 pKa = 4.36PCASDD365 pKa = 3.12TTPIVVPVILLRR377 pKa = 11.84NEE379 pKa = 4.58DD380 pKa = 3.73GDD382 pKa = 3.65GGNYY386 pKa = 9.83HH387 pKa = 7.05LVASTPVYY395 pKa = 9.46IGARR399 pKa = 11.84ARR401 pKa = 11.84RR402 pKa = 11.84SLDD405 pKa = 3.04MRR407 pKa = 11.84LISHH411 pKa = 7.47WNPCSAPKK419 pKa = 9.62PLCSMFEE426 pKa = 3.88LWCCVDD432 pKa = 3.51AVSAKK437 pKa = 10.38GADD440 pKa = 3.64VPSTRR445 pKa = 11.84DD446 pKa = 3.11DD447 pKa = 3.72SFILKK452 pKa = 9.26WIRR455 pKa = 11.84GRR457 pKa = 11.84VHH459 pKa = 6.81AALGRR464 pKa = 11.84WCRR467 pKa = 11.84VNRR470 pKa = 11.84ISSFTEE476 pKa = 3.7QEE478 pKa = 4.05GCEE481 pKa = 4.18TNNNSSSLITRR492 pKa = 11.84TTVCVFPSIPPNLEE506 pKa = 4.2TIDD509 pKa = 4.53HH510 pKa = 6.59ISFPVHH516 pKa = 5.77RR517 pKa = 11.84CVMSILFLSKK527 pKa = 10.92GSFPQSDD534 pKa = 3.86TTGSAVRR541 pKa = 11.84LHH543 pKa = 6.96RR544 pKa = 11.84MRR546 pKa = 11.84HH547 pKa = 3.92FQFARR552 pKa = 11.84KK553 pKa = 9.13LFSSLSDD560 pKa = 3.25TTATAWRR567 pKa = 11.84TMCHH571 pKa = 6.93DD572 pKa = 4.06DD573 pKa = 5.3EE574 pKa = 5.97KK575 pKa = 11.15NAWAFRR581 pKa = 11.84LCQSLGYY588 pKa = 9.27RR589 pKa = 11.84RR590 pKa = 11.84NVRR593 pKa = 11.84IIGEE597 pKa = 4.24EE598 pKa = 3.86EE599 pKa = 4.35RR600 pKa = 11.84EE601 pKa = 4.2TLLSLIGCPAFTDD614 pKa = 3.79VSGNDD619 pKa = 3.0ASFQISRR626 pKa = 11.84MARR629 pKa = 11.84LRR631 pKa = 11.84CSPSNAHH638 pKa = 5.87QRR640 pKa = 11.84VLLKK644 pKa = 9.96FAARR648 pKa = 11.84GGRR651 pKa = 11.84EE652 pKa = 3.83HH653 pKa = 6.27QRR655 pKa = 11.84EE656 pKa = 4.2GSALFVLKK664 pKa = 10.27PGSPAEE670 pKa = 4.05LFHH673 pKa = 8.03VGAVTSAPFFSMRR686 pKa = 11.84FGTRR690 pKa = 11.84VAYY693 pKa = 10.09AWVWDD698 pKa = 3.96DD699 pKa = 5.44AGWEE703 pKa = 4.09LVKK706 pKa = 10.71LYY708 pKa = 11.06NRR710 pKa = 11.84FALRR714 pKa = 11.84TVSRR718 pKa = 11.84EE719 pKa = 3.58EE720 pKa = 3.83TEE722 pKa = 4.17IFHH725 pKa = 6.98EE726 pKa = 5.2GPKK729 pKa = 9.6MGSATEE735 pKa = 3.98HH736 pKa = 6.37SGAGSGGNIYY746 pKa = 10.67FFLRR750 pKa = 11.84PLKK753 pKa = 10.11RR754 pKa = 11.84SHH756 pKa = 5.47VLYY759 pKa = 10.41RR760 pKa = 11.84FLRR763 pKa = 11.84ILPFIFLAVLLPLLLLRR780 pKa = 11.84AKK782 pKa = 9.75QVNKK786 pKa = 10.71SGITGMARR794 pKa = 11.84RR795 pKa = 11.84GG796 pKa = 3.34
MM1 pKa = 8.11DD2 pKa = 3.94VAQFLNVRR10 pKa = 11.84VEE12 pKa = 3.93EE13 pKa = 4.08LAAFSALLFRR23 pKa = 11.84KK24 pKa = 10.11RR25 pKa = 11.84PRR27 pKa = 11.84HH28 pKa = 4.86EE29 pKa = 3.92NEE31 pKa = 4.04GAFSCTARR39 pKa = 11.84SVVHH43 pKa = 5.44YY44 pKa = 9.69RR45 pKa = 11.84RR46 pKa = 11.84PSSANVPLSDD56 pKa = 3.94VMRR59 pKa = 11.84LAKK62 pKa = 10.1FRR64 pKa = 11.84SRR66 pKa = 11.84RR67 pKa = 11.84SKK69 pKa = 10.66RR70 pKa = 11.84FVRR73 pKa = 11.84LLAAEE78 pKa = 4.58EE79 pKa = 4.48PPPATPKK86 pKa = 10.57AVEE89 pKa = 4.08EE90 pKa = 4.32EE91 pKa = 4.0KK92 pKa = 11.32GRR94 pKa = 11.84MQKK97 pKa = 9.91KK98 pKa = 9.55RR99 pKa = 11.84RR100 pKa = 11.84LLTYY104 pKa = 10.15RR105 pKa = 11.84AWCLLLRR112 pKa = 11.84RR113 pKa = 11.84RR114 pKa = 11.84RR115 pKa = 11.84RR116 pKa = 11.84FLLRR120 pKa = 11.84RR121 pKa = 11.84RR122 pKa = 11.84ATTRR126 pKa = 11.84RR127 pKa = 11.84RR128 pKa = 11.84IRR130 pKa = 11.84RR131 pKa = 11.84RR132 pKa = 11.84IISLRR137 pKa = 11.84YY138 pKa = 9.86GKK140 pKa = 7.89TQLPFCNCGSAAGAPPPSFPPCLSTQSSSSGGGRR174 pKa = 11.84TAVLWLPSHH183 pKa = 6.17WRR185 pKa = 11.84FCRR188 pKa = 11.84RR189 pKa = 11.84FHH191 pKa = 6.73YY192 pKa = 10.69SVFKK196 pKa = 10.55TRR198 pKa = 11.84INGCSSSPDD207 pKa = 3.51DD208 pKa = 4.56DD209 pKa = 4.41FSRR212 pKa = 11.84LVNVRR217 pKa = 11.84IAVPTKK223 pKa = 8.66SQRR226 pKa = 11.84KK227 pKa = 4.49QHH229 pKa = 6.18RR230 pKa = 11.84VLQRR234 pKa = 11.84WAARR238 pKa = 11.84LPSCCALGKK247 pKa = 9.85PPGRR251 pKa = 11.84IKK253 pKa = 10.52SKK255 pKa = 10.33PMAVPPPLCFAAEE268 pKa = 4.0RR269 pKa = 11.84SHH271 pKa = 5.42MCVWAIQPRR280 pKa = 11.84EE281 pKa = 4.0NGHH284 pKa = 7.09LLTADD289 pKa = 3.41EE290 pKa = 4.61VMDD293 pKa = 3.3ILMIHH298 pKa = 6.09SCKK301 pKa = 10.4APPVALHH308 pKa = 5.93TRR310 pKa = 11.84DD311 pKa = 4.1LATMAQTSVGSVPHH325 pKa = 5.71EE326 pKa = 4.17KK327 pKa = 9.71CWRR330 pKa = 11.84RR331 pKa = 11.84TEE333 pKa = 3.98SAVFYY338 pKa = 10.44GYY340 pKa = 8.58MWSLGKK346 pKa = 10.58AMSKK350 pKa = 10.47SKK352 pKa = 9.94WLLRR356 pKa = 11.84GDD358 pKa = 4.25TEE360 pKa = 4.36PCASDD365 pKa = 3.12TTPIVVPVILLRR377 pKa = 11.84NEE379 pKa = 4.58DD380 pKa = 3.73GDD382 pKa = 3.65GGNYY386 pKa = 9.83HH387 pKa = 7.05LVASTPVYY395 pKa = 9.46IGARR399 pKa = 11.84ARR401 pKa = 11.84RR402 pKa = 11.84SLDD405 pKa = 3.04MRR407 pKa = 11.84LISHH411 pKa = 7.47WNPCSAPKK419 pKa = 9.62PLCSMFEE426 pKa = 3.88LWCCVDD432 pKa = 3.51AVSAKK437 pKa = 10.38GADD440 pKa = 3.64VPSTRR445 pKa = 11.84DD446 pKa = 3.11DD447 pKa = 3.72SFILKK452 pKa = 9.26WIRR455 pKa = 11.84GRR457 pKa = 11.84VHH459 pKa = 6.81AALGRR464 pKa = 11.84WCRR467 pKa = 11.84VNRR470 pKa = 11.84ISSFTEE476 pKa = 3.7QEE478 pKa = 4.05GCEE481 pKa = 4.18TNNNSSSLITRR492 pKa = 11.84TTVCVFPSIPPNLEE506 pKa = 4.2TIDD509 pKa = 4.53HH510 pKa = 6.59ISFPVHH516 pKa = 5.77RR517 pKa = 11.84CVMSILFLSKK527 pKa = 10.92GSFPQSDD534 pKa = 3.86TTGSAVRR541 pKa = 11.84LHH543 pKa = 6.96RR544 pKa = 11.84MRR546 pKa = 11.84HH547 pKa = 3.92FQFARR552 pKa = 11.84KK553 pKa = 9.13LFSSLSDD560 pKa = 3.25TTATAWRR567 pKa = 11.84TMCHH571 pKa = 6.93DD572 pKa = 4.06DD573 pKa = 5.3EE574 pKa = 5.97KK575 pKa = 11.15NAWAFRR581 pKa = 11.84LCQSLGYY588 pKa = 9.27RR589 pKa = 11.84RR590 pKa = 11.84NVRR593 pKa = 11.84IIGEE597 pKa = 4.24EE598 pKa = 3.86EE599 pKa = 4.35RR600 pKa = 11.84EE601 pKa = 4.2TLLSLIGCPAFTDD614 pKa = 3.79VSGNDD619 pKa = 3.0ASFQISRR626 pKa = 11.84MARR629 pKa = 11.84LRR631 pKa = 11.84CSPSNAHH638 pKa = 5.87QRR640 pKa = 11.84VLLKK644 pKa = 9.96FAARR648 pKa = 11.84GGRR651 pKa = 11.84EE652 pKa = 3.83HH653 pKa = 6.27QRR655 pKa = 11.84EE656 pKa = 4.2GSALFVLKK664 pKa = 10.27PGSPAEE670 pKa = 4.05LFHH673 pKa = 8.03VGAVTSAPFFSMRR686 pKa = 11.84FGTRR690 pKa = 11.84VAYY693 pKa = 10.09AWVWDD698 pKa = 3.96DD699 pKa = 5.44AGWEE703 pKa = 4.09LVKK706 pKa = 10.71LYY708 pKa = 11.06NRR710 pKa = 11.84FALRR714 pKa = 11.84TVSRR718 pKa = 11.84EE719 pKa = 3.58EE720 pKa = 3.83TEE722 pKa = 4.17IFHH725 pKa = 6.98EE726 pKa = 5.2GPKK729 pKa = 9.6MGSATEE735 pKa = 3.98HH736 pKa = 6.37SGAGSGGNIYY746 pKa = 10.67FFLRR750 pKa = 11.84PLKK753 pKa = 10.11RR754 pKa = 11.84SHH756 pKa = 5.47VLYY759 pKa = 10.41RR760 pKa = 11.84FLRR763 pKa = 11.84ILPFIFLAVLLPLLLLRR780 pKa = 11.84AKK782 pKa = 9.75QVNKK786 pKa = 10.71SGITGMARR794 pKa = 11.84RR795 pKa = 11.84GG796 pKa = 3.34
Molecular weight: 90.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4945837 |
10 |
5150 |
492.0 |
54.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.372 ± 0.024 | 2.091 ± 0.011 |
4.85 ± 0.014 | 6.977 ± 0.03 |
3.774 ± 0.014 | 6.736 ± 0.027 |
2.54 ± 0.01 | 3.866 ± 0.016 |
4.372 ± 0.018 | 9.699 ± 0.025 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.505 ± 0.008 | 3.65 ± 0.014 |
4.837 ± 0.019 | 3.728 ± 0.016 |
6.882 ± 0.022 | 7.988 ± 0.022 |
5.838 ± 0.015 | 7.626 ± 0.023 |
1.21 ± 0.008 | 2.458 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
