
Human adenovirus C serotype 6 (HAdV-6) (Human adenovirus 6)
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Preplasmiviricota; Tectiliviricetes; Rowavirales; Adenoviridae; Mastadenovirus; Human mastadenovirus C
Average proteome isoelectric point is 6.56
Get precalculated fractions of proteins
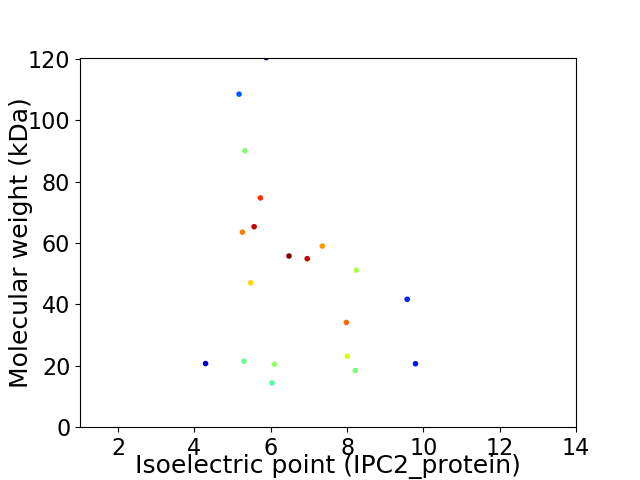
Virtual 2D-PAGE plot for 20 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E7DY82|E7DY82_ADE06 E1B 55 kDa protein OS=Human adenovirus C serotype 6 OX=10534 GN=E1B 55K PE=3 SV=1
MM1 pKa = 7.5RR2 pKa = 11.84HH3 pKa = 6.51IICHH7 pKa = 5.93GGVITEE13 pKa = 4.22EE14 pKa = 4.15MAASLLDD21 pKa = 3.6QLIEE25 pKa = 4.19EE26 pKa = 4.48VLADD30 pKa = 3.85NLPPPSHH37 pKa = 7.16FEE39 pKa = 3.8PPTLHH44 pKa = 6.78EE45 pKa = 5.5LYY47 pKa = 10.58DD48 pKa = 4.45LDD50 pKa = 3.62VTAPEE55 pKa = 4.89DD56 pKa = 3.83PNEE59 pKa = 4.13EE60 pKa = 3.85AVSQIFPEE68 pKa = 4.34SVMLAVQEE76 pKa = 4.97GIDD79 pKa = 4.38LFTFPPAPGSPEE91 pKa = 3.95PPHH94 pKa = 7.1LSRR97 pKa = 11.84QPEE100 pKa = 4.1QPEE103 pKa = 3.86QRR105 pKa = 11.84ALGPVSMPNLVPEE118 pKa = 4.78VIDD121 pKa = 3.89LTCHH125 pKa = 5.71EE126 pKa = 5.16AGFPPSDD133 pKa = 4.22DD134 pKa = 3.37EE135 pKa = 5.17DD136 pKa = 4.38EE137 pKa = 4.52EE138 pKa = 5.43GEE140 pKa = 4.43EE141 pKa = 4.08FVLDD145 pKa = 4.04YY146 pKa = 11.29VEE148 pKa = 5.48HH149 pKa = 7.23PGHH152 pKa = 6.6GCRR155 pKa = 11.84SCHH158 pKa = 4.33YY159 pKa = 9.82HH160 pKa = 6.49RR161 pKa = 11.84RR162 pKa = 11.84NTGDD166 pKa = 3.82PDD168 pKa = 3.74IMCSLCYY175 pKa = 9.84MRR177 pKa = 11.84TCGMFVYY184 pKa = 10.63SKK186 pKa = 11.39
MM1 pKa = 7.5RR2 pKa = 11.84HH3 pKa = 6.51IICHH7 pKa = 5.93GGVITEE13 pKa = 4.22EE14 pKa = 4.15MAASLLDD21 pKa = 3.6QLIEE25 pKa = 4.19EE26 pKa = 4.48VLADD30 pKa = 3.85NLPPPSHH37 pKa = 7.16FEE39 pKa = 3.8PPTLHH44 pKa = 6.78EE45 pKa = 5.5LYY47 pKa = 10.58DD48 pKa = 4.45LDD50 pKa = 3.62VTAPEE55 pKa = 4.89DD56 pKa = 3.83PNEE59 pKa = 4.13EE60 pKa = 3.85AVSQIFPEE68 pKa = 4.34SVMLAVQEE76 pKa = 4.97GIDD79 pKa = 4.38LFTFPPAPGSPEE91 pKa = 3.95PPHH94 pKa = 7.1LSRR97 pKa = 11.84QPEE100 pKa = 4.1QPEE103 pKa = 3.86QRR105 pKa = 11.84ALGPVSMPNLVPEE118 pKa = 4.78VIDD121 pKa = 3.89LTCHH125 pKa = 5.71EE126 pKa = 5.16AGFPPSDD133 pKa = 4.22DD134 pKa = 3.37EE135 pKa = 5.17DD136 pKa = 4.38EE137 pKa = 4.52EE138 pKa = 5.43GEE140 pKa = 4.43EE141 pKa = 4.08FVLDD145 pKa = 4.04YY146 pKa = 11.29VEE148 pKa = 5.48HH149 pKa = 7.23PGHH152 pKa = 6.6GCRR155 pKa = 11.84SCHH158 pKa = 4.33YY159 pKa = 9.82HH160 pKa = 6.49RR161 pKa = 11.84RR162 pKa = 11.84NTGDD166 pKa = 3.82PDD168 pKa = 3.74IMCSLCYY175 pKa = 9.84MRR177 pKa = 11.84TCGMFVYY184 pKa = 10.63SKK186 pKa = 11.39
Molecular weight: 20.73 kDa
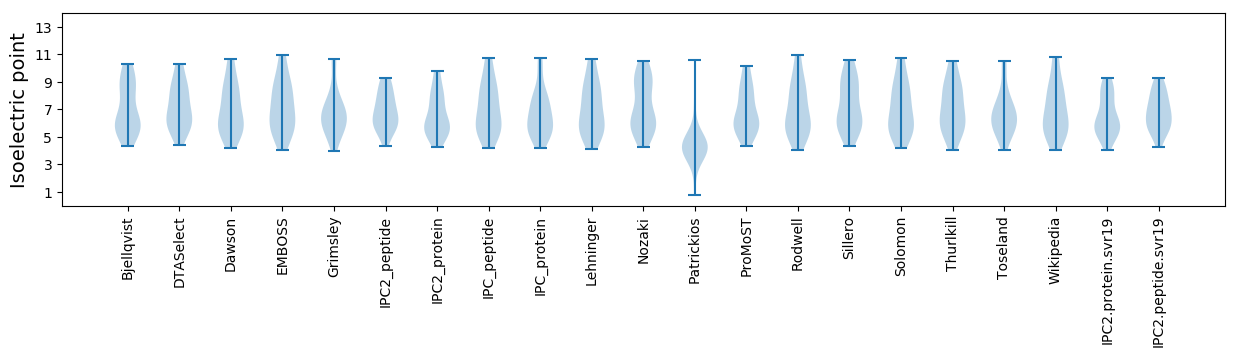
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E1U5P1|E1U5P1_ADE06 Isoform of Q3S8C4 Protease OS=Human adenovirus C serotype 6 OX=10534 GN=L3 PE=2 SV=1
MM1 pKa = 7.72SKK3 pKa = 10.63RR4 pKa = 11.84KK5 pKa = 9.53IKK7 pKa = 10.84EE8 pKa = 3.51EE9 pKa = 3.89MLQVIAPEE17 pKa = 4.21IYY19 pKa = 10.25GPPKK23 pKa = 10.38KK24 pKa = 10.29EE25 pKa = 3.84EE26 pKa = 3.69QDD28 pKa = 3.54YY29 pKa = 11.05KK30 pKa = 10.5PRR32 pKa = 11.84KK33 pKa = 8.67LKK35 pKa = 10.33RR36 pKa = 11.84VKK38 pKa = 10.18KK39 pKa = 10.1KK40 pKa = 10.88KK41 pKa = 10.23KK42 pKa = 10.35DD43 pKa = 3.74DD44 pKa = 4.83DD45 pKa = 4.92DD46 pKa = 6.89DD47 pKa = 4.46EE48 pKa = 7.23LDD50 pKa = 4.68DD51 pKa = 4.91EE52 pKa = 5.32VEE54 pKa = 4.32LLHH57 pKa = 6.36ATAPRR62 pKa = 11.84RR63 pKa = 11.84RR64 pKa = 11.84VQWKK68 pKa = 7.55GRR70 pKa = 11.84RR71 pKa = 11.84VRR73 pKa = 11.84RR74 pKa = 11.84VLRR77 pKa = 11.84PGTTVVFTPGEE88 pKa = 4.05RR89 pKa = 11.84STRR92 pKa = 11.84TYY94 pKa = 10.41KK95 pKa = 10.22RR96 pKa = 11.84VYY98 pKa = 10.52DD99 pKa = 3.86EE100 pKa = 4.9VYY102 pKa = 10.77GDD104 pKa = 4.06EE105 pKa = 5.51DD106 pKa = 4.34LLEE109 pKa = 4.21QANEE113 pKa = 3.98RR114 pKa = 11.84LGEE117 pKa = 3.73FAYY120 pKa = 10.17GKK122 pKa = 9.09RR123 pKa = 11.84HH124 pKa = 6.46KK125 pKa = 11.17DD126 pKa = 3.29MLALPLDD133 pKa = 4.36EE134 pKa = 5.85GNPTPSLKK142 pKa = 10.31PVTLQQVLPALAPSEE157 pKa = 4.33EE158 pKa = 3.88KK159 pKa = 10.48RR160 pKa = 11.84GLKK163 pKa = 10.08RR164 pKa = 11.84EE165 pKa = 4.22SGDD168 pKa = 3.66LAPTVQLMVPKK179 pKa = 10.19RR180 pKa = 11.84QRR182 pKa = 11.84LEE184 pKa = 4.07DD185 pKa = 3.69VLEE188 pKa = 4.36KK189 pKa = 9.08MTVEE193 pKa = 4.41PGLEE197 pKa = 3.84PEE199 pKa = 4.26VRR201 pKa = 11.84VRR203 pKa = 11.84PIKK206 pKa = 10.29QVAPGLGVQTVDD218 pKa = 3.28VQIPTTSSTSIATATEE234 pKa = 4.06GMEE237 pKa = 4.31TQTSPVASAVADD249 pKa = 3.75AAVQAAAAAASKK261 pKa = 9.84TSTEE265 pKa = 4.18VQTDD269 pKa = 2.66PWMFRR274 pKa = 11.84VSAPRR279 pKa = 11.84RR280 pKa = 11.84PRR282 pKa = 11.84RR283 pKa = 11.84SRR285 pKa = 11.84KK286 pKa = 9.48YY287 pKa = 8.94GAASALLPEE296 pKa = 4.69YY297 pKa = 10.54ALHH300 pKa = 6.8PSIAPTPGYY309 pKa = 9.62RR310 pKa = 11.84GYY312 pKa = 9.34TYY314 pKa = 10.47RR315 pKa = 11.84PRR317 pKa = 11.84RR318 pKa = 11.84RR319 pKa = 11.84ATTRR323 pKa = 11.84RR324 pKa = 11.84RR325 pKa = 11.84TTTGTRR331 pKa = 11.84RR332 pKa = 11.84RR333 pKa = 11.84RR334 pKa = 11.84RR335 pKa = 11.84RR336 pKa = 11.84RR337 pKa = 11.84QPVLAPISVRR347 pKa = 11.84RR348 pKa = 11.84VARR351 pKa = 11.84EE352 pKa = 3.67GGRR355 pKa = 11.84TLVLPTARR363 pKa = 11.84YY364 pKa = 8.7HH365 pKa = 6.0PSIVV369 pKa = 3.15
MM1 pKa = 7.72SKK3 pKa = 10.63RR4 pKa = 11.84KK5 pKa = 9.53IKK7 pKa = 10.84EE8 pKa = 3.51EE9 pKa = 3.89MLQVIAPEE17 pKa = 4.21IYY19 pKa = 10.25GPPKK23 pKa = 10.38KK24 pKa = 10.29EE25 pKa = 3.84EE26 pKa = 3.69QDD28 pKa = 3.54YY29 pKa = 11.05KK30 pKa = 10.5PRR32 pKa = 11.84KK33 pKa = 8.67LKK35 pKa = 10.33RR36 pKa = 11.84VKK38 pKa = 10.18KK39 pKa = 10.1KK40 pKa = 10.88KK41 pKa = 10.23KK42 pKa = 10.35DD43 pKa = 3.74DD44 pKa = 4.83DD45 pKa = 4.92DD46 pKa = 6.89DD47 pKa = 4.46EE48 pKa = 7.23LDD50 pKa = 4.68DD51 pKa = 4.91EE52 pKa = 5.32VEE54 pKa = 4.32LLHH57 pKa = 6.36ATAPRR62 pKa = 11.84RR63 pKa = 11.84RR64 pKa = 11.84VQWKK68 pKa = 7.55GRR70 pKa = 11.84RR71 pKa = 11.84VRR73 pKa = 11.84RR74 pKa = 11.84VLRR77 pKa = 11.84PGTTVVFTPGEE88 pKa = 4.05RR89 pKa = 11.84STRR92 pKa = 11.84TYY94 pKa = 10.41KK95 pKa = 10.22RR96 pKa = 11.84VYY98 pKa = 10.52DD99 pKa = 3.86EE100 pKa = 4.9VYY102 pKa = 10.77GDD104 pKa = 4.06EE105 pKa = 5.51DD106 pKa = 4.34LLEE109 pKa = 4.21QANEE113 pKa = 3.98RR114 pKa = 11.84LGEE117 pKa = 3.73FAYY120 pKa = 10.17GKK122 pKa = 9.09RR123 pKa = 11.84HH124 pKa = 6.46KK125 pKa = 11.17DD126 pKa = 3.29MLALPLDD133 pKa = 4.36EE134 pKa = 5.85GNPTPSLKK142 pKa = 10.31PVTLQQVLPALAPSEE157 pKa = 4.33EE158 pKa = 3.88KK159 pKa = 10.48RR160 pKa = 11.84GLKK163 pKa = 10.08RR164 pKa = 11.84EE165 pKa = 4.22SGDD168 pKa = 3.66LAPTVQLMVPKK179 pKa = 10.19RR180 pKa = 11.84QRR182 pKa = 11.84LEE184 pKa = 4.07DD185 pKa = 3.69VLEE188 pKa = 4.36KK189 pKa = 9.08MTVEE193 pKa = 4.41PGLEE197 pKa = 3.84PEE199 pKa = 4.26VRR201 pKa = 11.84VRR203 pKa = 11.84PIKK206 pKa = 10.29QVAPGLGVQTVDD218 pKa = 3.28VQIPTTSSTSIATATEE234 pKa = 4.06GMEE237 pKa = 4.31TQTSPVASAVADD249 pKa = 3.75AAVQAAAAAASKK261 pKa = 9.84TSTEE265 pKa = 4.18VQTDD269 pKa = 2.66PWMFRR274 pKa = 11.84VSAPRR279 pKa = 11.84RR280 pKa = 11.84PRR282 pKa = 11.84RR283 pKa = 11.84SRR285 pKa = 11.84KK286 pKa = 9.48YY287 pKa = 8.94GAASALLPEE296 pKa = 4.69YY297 pKa = 10.54ALHH300 pKa = 6.8PSIAPTPGYY309 pKa = 9.62RR310 pKa = 11.84GYY312 pKa = 9.34TYY314 pKa = 10.47RR315 pKa = 11.84PRR317 pKa = 11.84RR318 pKa = 11.84RR319 pKa = 11.84ATTRR323 pKa = 11.84RR324 pKa = 11.84RR325 pKa = 11.84TTTGTRR331 pKa = 11.84RR332 pKa = 11.84RR333 pKa = 11.84RR334 pKa = 11.84RR335 pKa = 11.84RR336 pKa = 11.84RR337 pKa = 11.84QPVLAPISVRR347 pKa = 11.84RR348 pKa = 11.84VARR351 pKa = 11.84EE352 pKa = 3.67GGRR355 pKa = 11.84TLVLPTARR363 pKa = 11.84YY364 pKa = 8.7HH365 pKa = 6.0PSIVV369 pKa = 3.15
Molecular weight: 41.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
8966 |
140 |
1056 |
448.3 |
50.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.231 ± 0.287 | 1.751 ± 0.329 |
5.365 ± 0.23 | 6.469 ± 0.504 |
3.703 ± 0.245 | 6.112 ± 0.244 |
2.331 ± 0.19 | 3.614 ± 0.18 |
3.759 ± 0.459 | 9.324 ± 0.408 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.61 ± 0.265 | 4.617 ± 0.498 |
6.781 ± 0.312 | 4.495 ± 0.28 |
7.45 ± 0.617 | 6.837 ± 0.413 |
5.934 ± 0.431 | 6.045 ± 0.264 |
1.26 ± 0.144 | 3.29 ± 0.283 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
