
Sclerotinia sclerotiorum megabirnavirus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Megabirnaviridae; Megabirnavirus; unclassified Megabirnavirus
Average proteome isoelectric point is 6.35
Get precalculated fractions of proteins
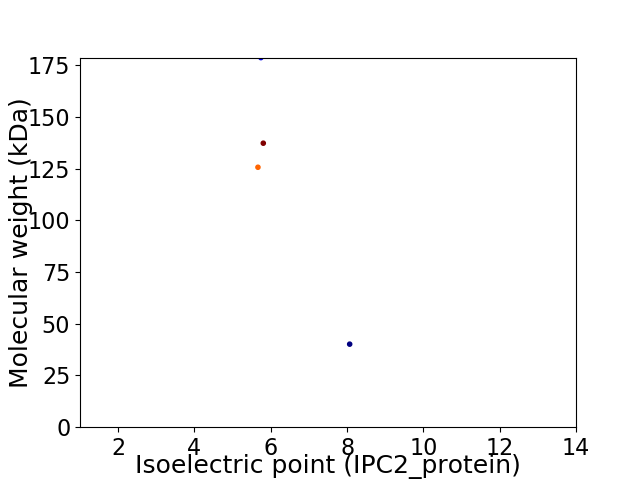
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0G3FL62|A0A0G3FL62_9VIRU Uncharacterized protein OS=Sclerotinia sclerotiorum megabirnavirus 1 OX=1661257 PE=4 SV=1
MM1 pKa = 7.51LANATALGEE10 pKa = 4.03QMLVIGDD17 pKa = 3.69EE18 pKa = 4.44LGRR21 pKa = 11.84EE22 pKa = 4.2SGDD25 pKa = 3.21AEE27 pKa = 4.03PRR29 pKa = 11.84GKK31 pKa = 9.11RR32 pKa = 11.84TLRR35 pKa = 11.84DD36 pKa = 3.45FPMLEE41 pKa = 4.07FSNAVHH47 pKa = 6.79AVMADD52 pKa = 3.47VEE54 pKa = 4.54LTSWDD59 pKa = 3.89AVCDD63 pKa = 3.57QHH65 pKa = 6.96TKK67 pKa = 8.68NTPQTLLVDD76 pKa = 4.09SPLLFSMAGQLLADD90 pKa = 4.28DD91 pKa = 5.06FARR94 pKa = 11.84AGVMIEE100 pKa = 3.78GRR102 pKa = 11.84QFLLLGRR109 pKa = 11.84NGHH112 pKa = 5.43TPVRR116 pKa = 11.84MDD118 pKa = 3.02TGTYY122 pKa = 10.81ASIKK126 pKa = 9.58TSLLRR131 pKa = 11.84HH132 pKa = 5.95ASPRR136 pKa = 11.84RR137 pKa = 11.84LRR139 pKa = 11.84AARR142 pKa = 11.84RR143 pKa = 11.84LFEE146 pKa = 4.84NVFAMNVPDD155 pKa = 4.99FLAVLEE161 pKa = 4.59AQHH164 pKa = 7.01DD165 pKa = 4.38CGPEE169 pKa = 3.62MDD171 pKa = 4.79SFFSMFSGDD180 pKa = 3.22GRR182 pKa = 11.84SLVVEE187 pKa = 4.75GAPTPCVGKK196 pKa = 9.94FLYY199 pKa = 10.03KK200 pKa = 10.47QLDD203 pKa = 3.48GGEE206 pKa = 4.16WVATDD211 pKa = 3.72VSAKK215 pKa = 8.76TASEE219 pKa = 3.92ALRR222 pKa = 11.84NAMVLRR228 pKa = 11.84KK229 pKa = 8.68LHH231 pKa = 7.04CDD233 pKa = 3.24GEE235 pKa = 4.65MGWEE239 pKa = 3.91QVLGLVEE246 pKa = 4.47RR247 pKa = 11.84SLISRR252 pKa = 11.84DD253 pKa = 3.11VHH255 pKa = 7.06LLSDD259 pKa = 4.78CIAYY263 pKa = 9.08LFPYY267 pKa = 8.31TEE269 pKa = 3.97RR270 pKa = 11.84SLYY273 pKa = 9.4QRR275 pKa = 11.84SRR277 pKa = 11.84MTFPLLSRR285 pKa = 11.84IILRR289 pKa = 11.84SIDD292 pKa = 3.87LGHH295 pKa = 5.76STEE298 pKa = 4.4GDD300 pKa = 3.07ILSLMRR306 pKa = 11.84HH307 pKa = 5.08MSGLGHH313 pKa = 5.81QASSAFFLYY322 pKa = 10.68SLWLPDD328 pKa = 3.27GAGRR332 pKa = 11.84HH333 pKa = 4.83VVDD336 pKa = 4.71FFCRR340 pKa = 11.84KK341 pKa = 7.89GWFNGGYY348 pKa = 9.06YY349 pKa = 10.41AVEE352 pKa = 3.97KK353 pKa = 10.33RR354 pKa = 11.84LKK356 pKa = 10.2ALHH359 pKa = 5.99SLARR363 pKa = 11.84RR364 pKa = 11.84CRR366 pKa = 11.84YY367 pKa = 9.04VPKK370 pKa = 10.19EE371 pKa = 3.47LRR373 pKa = 11.84GYY375 pKa = 8.9NVRR378 pKa = 11.84PTDD381 pKa = 3.74LMYY384 pKa = 9.99LTSVLGRR391 pKa = 11.84FYY393 pKa = 11.37LDD395 pKa = 4.11FAADD399 pKa = 3.84DD400 pKa = 4.09SAVSDD405 pKa = 3.26RR406 pKa = 11.84TEE408 pKa = 3.84YY409 pKa = 10.85YY410 pKa = 10.66GEE412 pKa = 4.1HH413 pKa = 6.64LSYY416 pKa = 11.31DD417 pKa = 3.87GVLGIYY423 pKa = 8.9STDD426 pKa = 2.94AHH428 pKa = 6.19KK429 pKa = 10.89QGMMRR434 pKa = 11.84FLLDD438 pKa = 3.55RR439 pKa = 11.84SRR441 pKa = 11.84VAGRR445 pKa = 11.84LWQQSEE451 pKa = 4.48VSSLGDD457 pKa = 2.74WMMRR461 pKa = 11.84YY462 pKa = 9.87VMYY465 pKa = 10.51GSSGSAGGHH474 pKa = 5.98SGDD477 pKa = 4.48EE478 pKa = 4.38FNVDD482 pKa = 2.95HH483 pKa = 6.69SVSKK487 pKa = 10.74RR488 pKa = 11.84LWLSNRR494 pKa = 11.84DD495 pKa = 3.06EE496 pKa = 5.24DD497 pKa = 3.93YY498 pKa = 11.23AAAYY502 pKa = 10.0VYY504 pKa = 9.83EE505 pKa = 4.51EE506 pKa = 4.12PAIASSVTVVKK517 pKa = 10.72RR518 pKa = 11.84EE519 pKa = 4.02AGKK522 pKa = 10.21LRR524 pKa = 11.84QLLPARR530 pKa = 11.84IPHH533 pKa = 6.93WLTEE537 pKa = 4.05SLLLSEE543 pKa = 4.85IEE545 pKa = 4.23SGILRR550 pKa = 11.84SMPLSLEE557 pKa = 3.9MSADD561 pKa = 3.29KK562 pKa = 11.28AMEE565 pKa = 4.33GVLQRR570 pKa = 11.84RR571 pKa = 11.84KK572 pKa = 10.96AMMSGEE578 pKa = 4.24TVACVDD584 pKa = 3.15WADD587 pKa = 3.9FNITHH592 pKa = 6.9TLSDD596 pKa = 3.23MAAYY600 pKa = 10.12FRR602 pKa = 11.84MLGKK606 pKa = 9.76QALEE610 pKa = 4.24TCTEE614 pKa = 3.87PNYY617 pKa = 10.48WNGISKK623 pKa = 10.11GQFLHH628 pKa = 7.44DD629 pKa = 4.03VAEE632 pKa = 4.71HH633 pKa = 6.66CATTLYY639 pKa = 10.85NLWIRR644 pKa = 11.84NGATADD650 pKa = 3.39SHH652 pKa = 7.16FEE654 pKa = 3.84HH655 pKa = 7.18AVRR658 pKa = 11.84GLWSGWRR665 pKa = 11.84STQFFNSSFNVAYY678 pKa = 10.8CEE680 pKa = 4.29TNFDD684 pKa = 3.43AMHH687 pKa = 7.24ALFQSPLPNRR697 pKa = 11.84ADD699 pKa = 3.36HH700 pKa = 7.13AGDD703 pKa = 4.58DD704 pKa = 4.4FFGTYY709 pKa = 10.26NDD711 pKa = 4.66PLDD714 pKa = 3.51AMRR717 pKa = 11.84FVLSMPLAQHH727 pKa = 6.67DD728 pKa = 4.45VNPTRR733 pKa = 11.84QLVDD737 pKa = 3.16GDD739 pKa = 3.49VGEE742 pKa = 4.47FLRR745 pKa = 11.84NEE747 pKa = 3.99YY748 pKa = 9.68MRR750 pKa = 11.84DD751 pKa = 3.86GIVHH755 pKa = 6.93GSFQRR760 pKa = 11.84SIGSFVGSDD769 pKa = 3.46LQSPEE774 pKa = 4.35LFSSITQAQGSNVRR788 pKa = 11.84RR789 pKa = 11.84GADD792 pKa = 3.05LSVIEE797 pKa = 4.58EE798 pKa = 3.87YY799 pKa = 10.38RR800 pKa = 11.84YY801 pKa = 8.81TIVQYY806 pKa = 9.73WATVYY811 pKa = 10.79SPEE814 pKa = 4.57SKK816 pKa = 9.72TSATPSVDD824 pKa = 3.54LCQLDD829 pKa = 4.25PTCGGLGCPRR839 pKa = 11.84YY840 pKa = 9.48GQQSTIATGSINTMPVQRR858 pKa = 11.84RR859 pKa = 11.84VPLDD863 pKa = 3.27VIMPRR868 pKa = 11.84NAAVLLQRR876 pKa = 11.84FKK878 pKa = 11.23QRR880 pKa = 11.84LWSYY884 pKa = 11.2NITSTNLEE892 pKa = 4.06GLRR895 pKa = 11.84DD896 pKa = 3.9DD897 pKa = 5.54VMIATYY903 pKa = 10.6GSDD906 pKa = 3.59FPAEE910 pKa = 3.73IKK912 pKa = 10.44KK913 pKa = 10.71RR914 pKa = 11.84LDD916 pKa = 3.13RR917 pKa = 11.84EE918 pKa = 4.05LRR920 pKa = 11.84EE921 pKa = 4.24TQVQWILEE929 pKa = 4.16ANKK932 pKa = 9.85RR933 pKa = 11.84VAVMRR938 pKa = 11.84KK939 pKa = 8.77AVLPAVPWWVDD950 pKa = 3.35DD951 pKa = 4.4YY952 pKa = 11.53IQTTIQRR959 pKa = 11.84VLANEE964 pKa = 4.0EE965 pKa = 4.15LDD967 pKa = 4.44GNSADD972 pKa = 4.13LNSMAMVVRR981 pKa = 11.84SMALAEE987 pKa = 4.21FSPLDD992 pKa = 3.39SALSHH997 pKa = 6.32LQTTAAQITGLPAIVKK1013 pKa = 9.77LSRR1016 pKa = 11.84GRR1018 pKa = 11.84VNPILSRR1025 pKa = 11.84MRR1027 pKa = 11.84VYY1029 pKa = 11.01LSDD1032 pKa = 3.76PMVEE1036 pKa = 4.0ALVEE1040 pKa = 4.27HH1041 pKa = 7.02RR1042 pKa = 11.84WNLPTSTGGVIPSEE1056 pKa = 4.08LRR1058 pKa = 11.84AVIYY1062 pKa = 10.68NVLNYY1067 pKa = 10.83ALYY1070 pKa = 8.81QVRR1073 pKa = 11.84YY1074 pKa = 8.73INQSFEE1080 pKa = 3.89SDD1082 pKa = 3.54ARR1084 pKa = 11.84WLTMYY1089 pKa = 8.34ATSVCQRR1096 pKa = 11.84LSNSWRR1102 pKa = 11.84QKK1104 pKa = 9.25CGTMFEE1110 pKa = 4.27IQDD1113 pKa = 4.13EE1114 pKa = 4.25ISYY1117 pKa = 11.14
MM1 pKa = 7.51LANATALGEE10 pKa = 4.03QMLVIGDD17 pKa = 3.69EE18 pKa = 4.44LGRR21 pKa = 11.84EE22 pKa = 4.2SGDD25 pKa = 3.21AEE27 pKa = 4.03PRR29 pKa = 11.84GKK31 pKa = 9.11RR32 pKa = 11.84TLRR35 pKa = 11.84DD36 pKa = 3.45FPMLEE41 pKa = 4.07FSNAVHH47 pKa = 6.79AVMADD52 pKa = 3.47VEE54 pKa = 4.54LTSWDD59 pKa = 3.89AVCDD63 pKa = 3.57QHH65 pKa = 6.96TKK67 pKa = 8.68NTPQTLLVDD76 pKa = 4.09SPLLFSMAGQLLADD90 pKa = 4.28DD91 pKa = 5.06FARR94 pKa = 11.84AGVMIEE100 pKa = 3.78GRR102 pKa = 11.84QFLLLGRR109 pKa = 11.84NGHH112 pKa = 5.43TPVRR116 pKa = 11.84MDD118 pKa = 3.02TGTYY122 pKa = 10.81ASIKK126 pKa = 9.58TSLLRR131 pKa = 11.84HH132 pKa = 5.95ASPRR136 pKa = 11.84RR137 pKa = 11.84LRR139 pKa = 11.84AARR142 pKa = 11.84RR143 pKa = 11.84LFEE146 pKa = 4.84NVFAMNVPDD155 pKa = 4.99FLAVLEE161 pKa = 4.59AQHH164 pKa = 7.01DD165 pKa = 4.38CGPEE169 pKa = 3.62MDD171 pKa = 4.79SFFSMFSGDD180 pKa = 3.22GRR182 pKa = 11.84SLVVEE187 pKa = 4.75GAPTPCVGKK196 pKa = 9.94FLYY199 pKa = 10.03KK200 pKa = 10.47QLDD203 pKa = 3.48GGEE206 pKa = 4.16WVATDD211 pKa = 3.72VSAKK215 pKa = 8.76TASEE219 pKa = 3.92ALRR222 pKa = 11.84NAMVLRR228 pKa = 11.84KK229 pKa = 8.68LHH231 pKa = 7.04CDD233 pKa = 3.24GEE235 pKa = 4.65MGWEE239 pKa = 3.91QVLGLVEE246 pKa = 4.47RR247 pKa = 11.84SLISRR252 pKa = 11.84DD253 pKa = 3.11VHH255 pKa = 7.06LLSDD259 pKa = 4.78CIAYY263 pKa = 9.08LFPYY267 pKa = 8.31TEE269 pKa = 3.97RR270 pKa = 11.84SLYY273 pKa = 9.4QRR275 pKa = 11.84SRR277 pKa = 11.84MTFPLLSRR285 pKa = 11.84IILRR289 pKa = 11.84SIDD292 pKa = 3.87LGHH295 pKa = 5.76STEE298 pKa = 4.4GDD300 pKa = 3.07ILSLMRR306 pKa = 11.84HH307 pKa = 5.08MSGLGHH313 pKa = 5.81QASSAFFLYY322 pKa = 10.68SLWLPDD328 pKa = 3.27GAGRR332 pKa = 11.84HH333 pKa = 4.83VVDD336 pKa = 4.71FFCRR340 pKa = 11.84KK341 pKa = 7.89GWFNGGYY348 pKa = 9.06YY349 pKa = 10.41AVEE352 pKa = 3.97KK353 pKa = 10.33RR354 pKa = 11.84LKK356 pKa = 10.2ALHH359 pKa = 5.99SLARR363 pKa = 11.84RR364 pKa = 11.84CRR366 pKa = 11.84YY367 pKa = 9.04VPKK370 pKa = 10.19EE371 pKa = 3.47LRR373 pKa = 11.84GYY375 pKa = 8.9NVRR378 pKa = 11.84PTDD381 pKa = 3.74LMYY384 pKa = 9.99LTSVLGRR391 pKa = 11.84FYY393 pKa = 11.37LDD395 pKa = 4.11FAADD399 pKa = 3.84DD400 pKa = 4.09SAVSDD405 pKa = 3.26RR406 pKa = 11.84TEE408 pKa = 3.84YY409 pKa = 10.85YY410 pKa = 10.66GEE412 pKa = 4.1HH413 pKa = 6.64LSYY416 pKa = 11.31DD417 pKa = 3.87GVLGIYY423 pKa = 8.9STDD426 pKa = 2.94AHH428 pKa = 6.19KK429 pKa = 10.89QGMMRR434 pKa = 11.84FLLDD438 pKa = 3.55RR439 pKa = 11.84SRR441 pKa = 11.84VAGRR445 pKa = 11.84LWQQSEE451 pKa = 4.48VSSLGDD457 pKa = 2.74WMMRR461 pKa = 11.84YY462 pKa = 9.87VMYY465 pKa = 10.51GSSGSAGGHH474 pKa = 5.98SGDD477 pKa = 4.48EE478 pKa = 4.38FNVDD482 pKa = 2.95HH483 pKa = 6.69SVSKK487 pKa = 10.74RR488 pKa = 11.84LWLSNRR494 pKa = 11.84DD495 pKa = 3.06EE496 pKa = 5.24DD497 pKa = 3.93YY498 pKa = 11.23AAAYY502 pKa = 10.0VYY504 pKa = 9.83EE505 pKa = 4.51EE506 pKa = 4.12PAIASSVTVVKK517 pKa = 10.72RR518 pKa = 11.84EE519 pKa = 4.02AGKK522 pKa = 10.21LRR524 pKa = 11.84QLLPARR530 pKa = 11.84IPHH533 pKa = 6.93WLTEE537 pKa = 4.05SLLLSEE543 pKa = 4.85IEE545 pKa = 4.23SGILRR550 pKa = 11.84SMPLSLEE557 pKa = 3.9MSADD561 pKa = 3.29KK562 pKa = 11.28AMEE565 pKa = 4.33GVLQRR570 pKa = 11.84RR571 pKa = 11.84KK572 pKa = 10.96AMMSGEE578 pKa = 4.24TVACVDD584 pKa = 3.15WADD587 pKa = 3.9FNITHH592 pKa = 6.9TLSDD596 pKa = 3.23MAAYY600 pKa = 10.12FRR602 pKa = 11.84MLGKK606 pKa = 9.76QALEE610 pKa = 4.24TCTEE614 pKa = 3.87PNYY617 pKa = 10.48WNGISKK623 pKa = 10.11GQFLHH628 pKa = 7.44DD629 pKa = 4.03VAEE632 pKa = 4.71HH633 pKa = 6.66CATTLYY639 pKa = 10.85NLWIRR644 pKa = 11.84NGATADD650 pKa = 3.39SHH652 pKa = 7.16FEE654 pKa = 3.84HH655 pKa = 7.18AVRR658 pKa = 11.84GLWSGWRR665 pKa = 11.84STQFFNSSFNVAYY678 pKa = 10.8CEE680 pKa = 4.29TNFDD684 pKa = 3.43AMHH687 pKa = 7.24ALFQSPLPNRR697 pKa = 11.84ADD699 pKa = 3.36HH700 pKa = 7.13AGDD703 pKa = 4.58DD704 pKa = 4.4FFGTYY709 pKa = 10.26NDD711 pKa = 4.66PLDD714 pKa = 3.51AMRR717 pKa = 11.84FVLSMPLAQHH727 pKa = 6.67DD728 pKa = 4.45VNPTRR733 pKa = 11.84QLVDD737 pKa = 3.16GDD739 pKa = 3.49VGEE742 pKa = 4.47FLRR745 pKa = 11.84NEE747 pKa = 3.99YY748 pKa = 9.68MRR750 pKa = 11.84DD751 pKa = 3.86GIVHH755 pKa = 6.93GSFQRR760 pKa = 11.84SIGSFVGSDD769 pKa = 3.46LQSPEE774 pKa = 4.35LFSSITQAQGSNVRR788 pKa = 11.84RR789 pKa = 11.84GADD792 pKa = 3.05LSVIEE797 pKa = 4.58EE798 pKa = 3.87YY799 pKa = 10.38RR800 pKa = 11.84YY801 pKa = 8.81TIVQYY806 pKa = 9.73WATVYY811 pKa = 10.79SPEE814 pKa = 4.57SKK816 pKa = 9.72TSATPSVDD824 pKa = 3.54LCQLDD829 pKa = 4.25PTCGGLGCPRR839 pKa = 11.84YY840 pKa = 9.48GQQSTIATGSINTMPVQRR858 pKa = 11.84RR859 pKa = 11.84VPLDD863 pKa = 3.27VIMPRR868 pKa = 11.84NAAVLLQRR876 pKa = 11.84FKK878 pKa = 11.23QRR880 pKa = 11.84LWSYY884 pKa = 11.2NITSTNLEE892 pKa = 4.06GLRR895 pKa = 11.84DD896 pKa = 3.9DD897 pKa = 5.54VMIATYY903 pKa = 10.6GSDD906 pKa = 3.59FPAEE910 pKa = 3.73IKK912 pKa = 10.44KK913 pKa = 10.71RR914 pKa = 11.84LDD916 pKa = 3.13RR917 pKa = 11.84EE918 pKa = 4.05LRR920 pKa = 11.84EE921 pKa = 4.24TQVQWILEE929 pKa = 4.16ANKK932 pKa = 9.85RR933 pKa = 11.84VAVMRR938 pKa = 11.84KK939 pKa = 8.77AVLPAVPWWVDD950 pKa = 3.35DD951 pKa = 4.4YY952 pKa = 11.53IQTTIQRR959 pKa = 11.84VLANEE964 pKa = 4.0EE965 pKa = 4.15LDD967 pKa = 4.44GNSADD972 pKa = 4.13LNSMAMVVRR981 pKa = 11.84SMALAEE987 pKa = 4.21FSPLDD992 pKa = 3.39SALSHH997 pKa = 6.32LQTTAAQITGLPAIVKK1013 pKa = 9.77LSRR1016 pKa = 11.84GRR1018 pKa = 11.84VNPILSRR1025 pKa = 11.84MRR1027 pKa = 11.84VYY1029 pKa = 11.01LSDD1032 pKa = 3.76PMVEE1036 pKa = 4.0ALVEE1040 pKa = 4.27HH1041 pKa = 7.02RR1042 pKa = 11.84WNLPTSTGGVIPSEE1056 pKa = 4.08LRR1058 pKa = 11.84AVIYY1062 pKa = 10.68NVLNYY1067 pKa = 10.83ALYY1070 pKa = 8.81QVRR1073 pKa = 11.84YY1074 pKa = 8.73INQSFEE1080 pKa = 3.89SDD1082 pKa = 3.54ARR1084 pKa = 11.84WLTMYY1089 pKa = 8.34ATSVCQRR1096 pKa = 11.84LSNSWRR1102 pKa = 11.84QKK1104 pKa = 9.25CGTMFEE1110 pKa = 4.27IQDD1113 pKa = 4.13EE1114 pKa = 4.25ISYY1117 pKa = 11.14
Molecular weight: 125.6 kDa
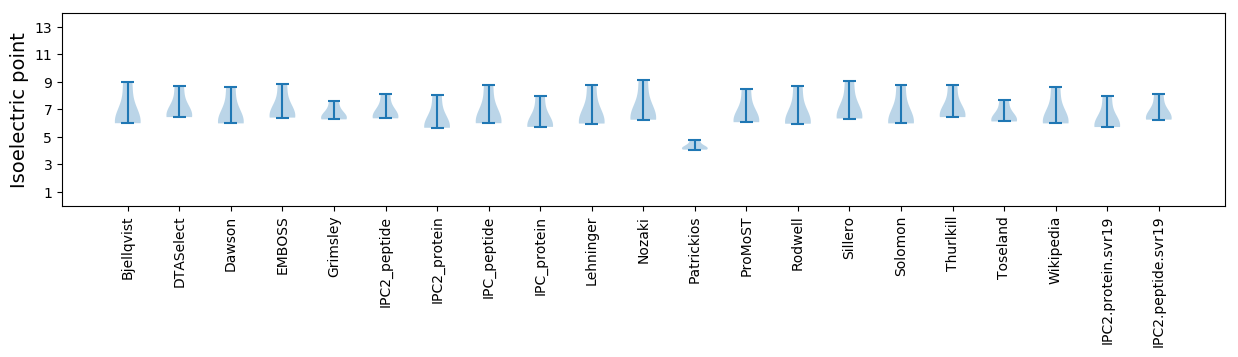
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0G3FL62|A0A0G3FL62_9VIRU Uncharacterized protein OS=Sclerotinia sclerotiorum megabirnavirus 1 OX=1661257 PE=4 SV=1
MM1 pKa = 7.31VKK3 pKa = 10.29RR4 pKa = 11.84EE5 pKa = 4.26GFRR8 pKa = 11.84ITYY11 pKa = 7.63VWRR14 pKa = 11.84TYY16 pKa = 10.68EE17 pKa = 3.93YY18 pKa = 11.31GRR20 pKa = 11.84LALLYY25 pKa = 9.93RR26 pKa = 11.84PQDD29 pKa = 3.03RR30 pKa = 11.84GRR32 pKa = 11.84SVLAQLIVRR41 pKa = 11.84NGPALLRR48 pKa = 11.84DD49 pKa = 3.83MRR51 pKa = 11.84SGTTRR56 pKa = 11.84NSLRR60 pKa = 11.84VMVCISALVSLLARR74 pKa = 11.84ARR76 pKa = 11.84TTLVKK81 pKa = 10.65VKK83 pKa = 10.06QAEE86 pKa = 4.18LASAEE91 pKa = 3.89QRR93 pKa = 11.84DD94 pKa = 3.85RR95 pKa = 11.84EE96 pKa = 3.72RR97 pKa = 11.84SYY99 pKa = 9.87ILRR102 pKa = 11.84RR103 pKa = 11.84NGHH106 pKa = 5.64CRR108 pKa = 11.84AVIRR112 pKa = 11.84YY113 pKa = 8.7AIARR117 pKa = 11.84RR118 pKa = 11.84DD119 pKa = 3.57EE120 pKa = 4.1QVRR123 pKa = 11.84TEE125 pKa = 4.01AVRR128 pKa = 11.84AEE130 pKa = 4.31TFAEE134 pKa = 3.76QRR136 pKa = 11.84LISEE140 pKa = 4.6HH141 pKa = 5.88SAIVDD146 pKa = 3.97LVAQLLAEE154 pKa = 4.08EE155 pKa = 4.83AEE157 pKa = 4.54PLFEE161 pKa = 3.88AHH163 pKa = 6.97KK164 pKa = 10.11AAEE167 pKa = 4.11VSPTLVRR174 pKa = 11.84EE175 pKa = 4.16DD176 pKa = 3.51QAPEE180 pKa = 3.84NVPEE184 pKa = 4.13EE185 pKa = 4.17EE186 pKa = 4.57GEE188 pKa = 3.87ISDD191 pKa = 4.35FVFGVAAVWRR201 pKa = 11.84AINIRR206 pKa = 11.84AWILRR211 pKa = 11.84GQYY214 pKa = 10.32NYY216 pKa = 8.49RR217 pKa = 11.84TRR219 pKa = 11.84KK220 pKa = 9.22LAQQRR225 pKa = 11.84LAEE228 pKa = 4.34MNDD231 pKa = 2.99VAARR235 pKa = 11.84ASDD238 pKa = 3.92DD239 pKa = 3.66VGDD242 pKa = 3.78TSGRR246 pKa = 11.84QPSEE250 pKa = 3.34AATFGHH256 pKa = 6.69HH257 pKa = 6.35FSCFPALGICNCDD270 pKa = 2.99AHH272 pKa = 6.88AVSAGAPAVEE282 pKa = 4.3VMTDD286 pKa = 3.2EE287 pKa = 4.42QVCKK291 pKa = 10.22KK292 pKa = 10.69CEE294 pKa = 4.04IITAQVFEE302 pKa = 4.7RR303 pKa = 11.84NGRR306 pKa = 11.84ANVSIKK312 pKa = 10.33VVPPMFARR320 pKa = 11.84AAFEE324 pKa = 4.71ADD326 pKa = 3.55GSDD329 pKa = 3.54LRR331 pKa = 11.84IWNIAVRR338 pKa = 11.84RR339 pKa = 11.84AGYY342 pKa = 10.27EE343 pKa = 3.61EE344 pKa = 6.13LIPKK348 pKa = 8.55MRR350 pKa = 11.84TILANSTGG358 pKa = 3.37
MM1 pKa = 7.31VKK3 pKa = 10.29RR4 pKa = 11.84EE5 pKa = 4.26GFRR8 pKa = 11.84ITYY11 pKa = 7.63VWRR14 pKa = 11.84TYY16 pKa = 10.68EE17 pKa = 3.93YY18 pKa = 11.31GRR20 pKa = 11.84LALLYY25 pKa = 9.93RR26 pKa = 11.84PQDD29 pKa = 3.03RR30 pKa = 11.84GRR32 pKa = 11.84SVLAQLIVRR41 pKa = 11.84NGPALLRR48 pKa = 11.84DD49 pKa = 3.83MRR51 pKa = 11.84SGTTRR56 pKa = 11.84NSLRR60 pKa = 11.84VMVCISALVSLLARR74 pKa = 11.84ARR76 pKa = 11.84TTLVKK81 pKa = 10.65VKK83 pKa = 10.06QAEE86 pKa = 4.18LASAEE91 pKa = 3.89QRR93 pKa = 11.84DD94 pKa = 3.85RR95 pKa = 11.84EE96 pKa = 3.72RR97 pKa = 11.84SYY99 pKa = 9.87ILRR102 pKa = 11.84RR103 pKa = 11.84NGHH106 pKa = 5.64CRR108 pKa = 11.84AVIRR112 pKa = 11.84YY113 pKa = 8.7AIARR117 pKa = 11.84RR118 pKa = 11.84DD119 pKa = 3.57EE120 pKa = 4.1QVRR123 pKa = 11.84TEE125 pKa = 4.01AVRR128 pKa = 11.84AEE130 pKa = 4.31TFAEE134 pKa = 3.76QRR136 pKa = 11.84LISEE140 pKa = 4.6HH141 pKa = 5.88SAIVDD146 pKa = 3.97LVAQLLAEE154 pKa = 4.08EE155 pKa = 4.83AEE157 pKa = 4.54PLFEE161 pKa = 3.88AHH163 pKa = 6.97KK164 pKa = 10.11AAEE167 pKa = 4.11VSPTLVRR174 pKa = 11.84EE175 pKa = 4.16DD176 pKa = 3.51QAPEE180 pKa = 3.84NVPEE184 pKa = 4.13EE185 pKa = 4.17EE186 pKa = 4.57GEE188 pKa = 3.87ISDD191 pKa = 4.35FVFGVAAVWRR201 pKa = 11.84AINIRR206 pKa = 11.84AWILRR211 pKa = 11.84GQYY214 pKa = 10.32NYY216 pKa = 8.49RR217 pKa = 11.84TRR219 pKa = 11.84KK220 pKa = 9.22LAQQRR225 pKa = 11.84LAEE228 pKa = 4.34MNDD231 pKa = 2.99VAARR235 pKa = 11.84ASDD238 pKa = 3.92DD239 pKa = 3.66VGDD242 pKa = 3.78TSGRR246 pKa = 11.84QPSEE250 pKa = 3.34AATFGHH256 pKa = 6.69HH257 pKa = 6.35FSCFPALGICNCDD270 pKa = 2.99AHH272 pKa = 6.88AVSAGAPAVEE282 pKa = 4.3VMTDD286 pKa = 3.2EE287 pKa = 4.42QVCKK291 pKa = 10.22KK292 pKa = 10.69CEE294 pKa = 4.04IITAQVFEE302 pKa = 4.7RR303 pKa = 11.84NGRR306 pKa = 11.84ANVSIKK312 pKa = 10.33VVPPMFARR320 pKa = 11.84AAFEE324 pKa = 4.71ADD326 pKa = 3.55GSDD329 pKa = 3.54LRR331 pKa = 11.84IWNIAVRR338 pKa = 11.84RR339 pKa = 11.84AGYY342 pKa = 10.27EE343 pKa = 3.61EE344 pKa = 6.13LIPKK348 pKa = 8.55MRR350 pKa = 11.84TILANSTGG358 pKa = 3.37
Molecular weight: 40.13 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4352 |
358 |
1626 |
1088.0 |
120.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.754 ± 0.862 | 1.425 ± 0.243 |
6.089 ± 0.284 | 5.193 ± 0.369 |
3.47 ± 0.376 | 7.652 ± 0.472 |
2.528 ± 0.115 | 3.608 ± 0.211 |
2.459 ± 0.072 | 9.421 ± 0.754 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.056 ± 0.413 | 4.435 ± 0.611 |
3.814 ± 0.154 | 3.814 ± 0.029 |
7.79 ± 0.544 | 6.434 ± 0.81 |
5.79 ± 0.317 | 7.399 ± 0.357 |
1.769 ± 0.132 | 3.102 ± 0.314 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
