
Nocardioides sp. BN140041
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Propionibacteriales; Nocardioidaceae; Nocardioides
Average proteome isoelectric point is 5.85
Get precalculated fractions of proteins
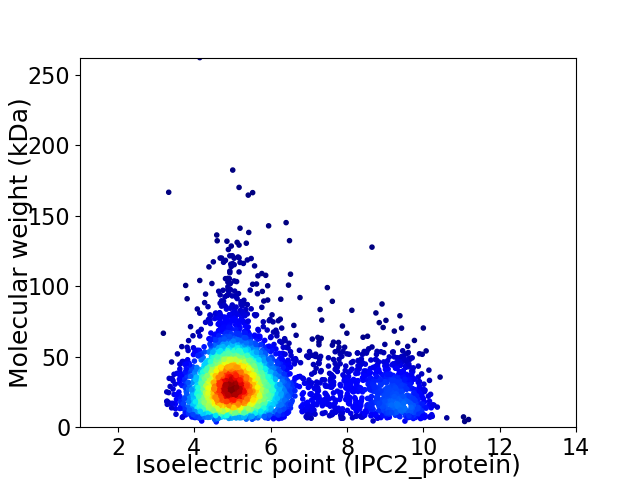
Virtual 2D-PAGE plot for 4050 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5B1M656|A0A5B1M656_9ACTN Beta-glucosidase OS=Nocardioides sp. BN140041 OX=2607659 GN=F0U47_11055 PE=3 SV=1
MM1 pKa = 7.61KK2 pKa = 9.21NTTRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84TVCGISLLSALSLAVAACGNGSDD32 pKa = 3.96EE33 pKa = 5.49AGDD36 pKa = 3.87EE37 pKa = 4.25NEE39 pKa = 4.34LTFVNYY45 pKa = 10.61GGDD48 pKa = 3.31GMEE51 pKa = 4.09AAKK54 pKa = 10.66AGWLEE59 pKa = 4.26PYY61 pKa = 9.19TEE63 pKa = 4.19KK64 pKa = 10.49TGVTFEE70 pKa = 4.23TDD72 pKa = 3.24EE73 pKa = 5.19PSDD76 pKa = 3.62QAKK79 pKa = 9.51IKK81 pKa = 11.1AMVASGKK88 pKa = 7.35VTWDD92 pKa = 3.27VVDD95 pKa = 4.35IDD97 pKa = 4.13AASAGPEE104 pKa = 4.03CGTLYY109 pKa = 10.67EE110 pKa = 4.56EE111 pKa = 4.96RR112 pKa = 11.84PSDD115 pKa = 3.69FDD117 pKa = 3.46TSEE120 pKa = 3.99LMEE123 pKa = 4.52NEE125 pKa = 3.79ITDD128 pKa = 3.62DD129 pKa = 4.11CMIPIINQTVALVYY143 pKa = 10.36NAEE146 pKa = 4.44LYY148 pKa = 10.94GDD150 pKa = 4.25NPPTSITDD158 pKa = 3.75FFNPEE163 pKa = 3.77FEE165 pKa = 4.33GQRR168 pKa = 11.84ITFSYY173 pKa = 7.3WTGSAEE179 pKa = 4.15PLALAAGIPAEE190 pKa = 4.4DD191 pKa = 4.69VYY193 pKa = 11.19PIDD196 pKa = 3.46WDD198 pKa = 3.88EE199 pKa = 4.74LKK201 pKa = 11.0SAVDD205 pKa = 3.58SLGSDD210 pKa = 3.47LTFQSTLDD218 pKa = 3.45QQVQSLEE225 pKa = 4.07SGNFGMCLCYY235 pKa = 9.89TGRR238 pKa = 11.84SALAAQNGADD248 pKa = 4.66IGVVWDD254 pKa = 4.28KK255 pKa = 11.58VWVGWDD261 pKa = 3.11GLYY264 pKa = 10.31VVKK267 pKa = 10.68DD268 pKa = 3.65SPRR271 pKa = 11.84EE272 pKa = 4.01DD273 pKa = 3.51IAWDD277 pKa = 4.17FVQYY281 pKa = 10.91LATSEE286 pKa = 4.31GQNGYY291 pKa = 9.39YY292 pKa = 10.02EE293 pKa = 4.66HH294 pKa = 7.26LPYY297 pKa = 10.89SPTTTGAPPDD307 pKa = 3.56VPEE310 pKa = 4.02EE311 pKa = 4.07FKK313 pKa = 11.19PFMLSFNEE321 pKa = 4.28GKK323 pKa = 9.9VQEE326 pKa = 4.35TSYY329 pKa = 11.39FDD331 pKa = 2.96AGYY334 pKa = 9.52WGDD337 pKa = 3.75QGATAADD344 pKa = 3.69EE345 pKa = 4.42WTALTAGG352 pKa = 3.85
MM1 pKa = 7.61KK2 pKa = 9.21NTTRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84TVCGISLLSALSLAVAACGNGSDD32 pKa = 3.96EE33 pKa = 5.49AGDD36 pKa = 3.87EE37 pKa = 4.25NEE39 pKa = 4.34LTFVNYY45 pKa = 10.61GGDD48 pKa = 3.31GMEE51 pKa = 4.09AAKK54 pKa = 10.66AGWLEE59 pKa = 4.26PYY61 pKa = 9.19TEE63 pKa = 4.19KK64 pKa = 10.49TGVTFEE70 pKa = 4.23TDD72 pKa = 3.24EE73 pKa = 5.19PSDD76 pKa = 3.62QAKK79 pKa = 9.51IKK81 pKa = 11.1AMVASGKK88 pKa = 7.35VTWDD92 pKa = 3.27VVDD95 pKa = 4.35IDD97 pKa = 4.13AASAGPEE104 pKa = 4.03CGTLYY109 pKa = 10.67EE110 pKa = 4.56EE111 pKa = 4.96RR112 pKa = 11.84PSDD115 pKa = 3.69FDD117 pKa = 3.46TSEE120 pKa = 3.99LMEE123 pKa = 4.52NEE125 pKa = 3.79ITDD128 pKa = 3.62DD129 pKa = 4.11CMIPIINQTVALVYY143 pKa = 10.36NAEE146 pKa = 4.44LYY148 pKa = 10.94GDD150 pKa = 4.25NPPTSITDD158 pKa = 3.75FFNPEE163 pKa = 3.77FEE165 pKa = 4.33GQRR168 pKa = 11.84ITFSYY173 pKa = 7.3WTGSAEE179 pKa = 4.15PLALAAGIPAEE190 pKa = 4.4DD191 pKa = 4.69VYY193 pKa = 11.19PIDD196 pKa = 3.46WDD198 pKa = 3.88EE199 pKa = 4.74LKK201 pKa = 11.0SAVDD205 pKa = 3.58SLGSDD210 pKa = 3.47LTFQSTLDD218 pKa = 3.45QQVQSLEE225 pKa = 4.07SGNFGMCLCYY235 pKa = 9.89TGRR238 pKa = 11.84SALAAQNGADD248 pKa = 4.66IGVVWDD254 pKa = 4.28KK255 pKa = 11.58VWVGWDD261 pKa = 3.11GLYY264 pKa = 10.31VVKK267 pKa = 10.68DD268 pKa = 3.65SPRR271 pKa = 11.84EE272 pKa = 4.01DD273 pKa = 3.51IAWDD277 pKa = 4.17FVQYY281 pKa = 10.91LATSEE286 pKa = 4.31GQNGYY291 pKa = 9.39YY292 pKa = 10.02EE293 pKa = 4.66HH294 pKa = 7.26LPYY297 pKa = 10.89SPTTTGAPPDD307 pKa = 3.56VPEE310 pKa = 4.02EE311 pKa = 4.07FKK313 pKa = 11.19PFMLSFNEE321 pKa = 4.28GKK323 pKa = 9.9VQEE326 pKa = 4.35TSYY329 pKa = 11.39FDD331 pKa = 2.96AGYY334 pKa = 9.52WGDD337 pKa = 3.75QGATAADD344 pKa = 3.69EE345 pKa = 4.42WTALTAGG352 pKa = 3.85
Molecular weight: 38.29 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5B1LZD7|A0A5B1LZD7_9ACTN Probable membrane transporter protein OS=Nocardioides sp. BN140041 OX=2607659 GN=F0U47_17060 PE=3 SV=1
MM1 pKa = 7.47RR2 pKa = 11.84RR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84ARR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84PPRR12 pKa = 11.84PPARR16 pKa = 11.84WTPPRR21 pKa = 11.84PWRR24 pKa = 11.84APRR27 pKa = 11.84PPSRR31 pKa = 11.84RR32 pKa = 11.84CPRR35 pKa = 11.84SGRR38 pKa = 11.84PARR41 pKa = 11.84PWSPTPTRR49 pKa = 11.84RR50 pKa = 11.84SRR52 pKa = 11.84PGRR55 pKa = 11.84RR56 pKa = 11.84GGRR59 pKa = 11.84GCC61 pKa = 4.61
MM1 pKa = 7.47RR2 pKa = 11.84RR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84ARR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84PPRR12 pKa = 11.84PPARR16 pKa = 11.84WTPPRR21 pKa = 11.84PWRR24 pKa = 11.84APRR27 pKa = 11.84PPSRR31 pKa = 11.84RR32 pKa = 11.84CPRR35 pKa = 11.84SGRR38 pKa = 11.84PARR41 pKa = 11.84PWSPTPTRR49 pKa = 11.84RR50 pKa = 11.84SRR52 pKa = 11.84PGRR55 pKa = 11.84RR56 pKa = 11.84GGRR59 pKa = 11.84GCC61 pKa = 4.61
Molecular weight: 7.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1263282 |
33 |
2527 |
311.9 |
33.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.816 ± 0.049 | 0.772 ± 0.011 |
6.712 ± 0.037 | 6.086 ± 0.035 |
2.817 ± 0.018 | 9.162 ± 0.036 |
2.165 ± 0.02 | 3.596 ± 0.024 |
2.075 ± 0.027 | 10.105 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.804 ± 0.016 | 1.808 ± 0.022 |
5.673 ± 0.03 | 2.713 ± 0.021 |
7.696 ± 0.046 | 5.056 ± 0.022 |
5.961 ± 0.035 | 9.399 ± 0.038 |
1.527 ± 0.016 | 2.056 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
