
Marinovum algicola DG 898
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Marinovum; Marinovum algicola
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
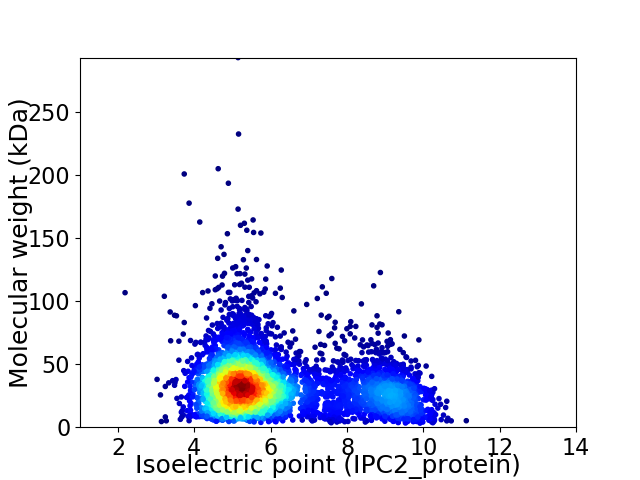
Virtual 2D-PAGE plot for 4976 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
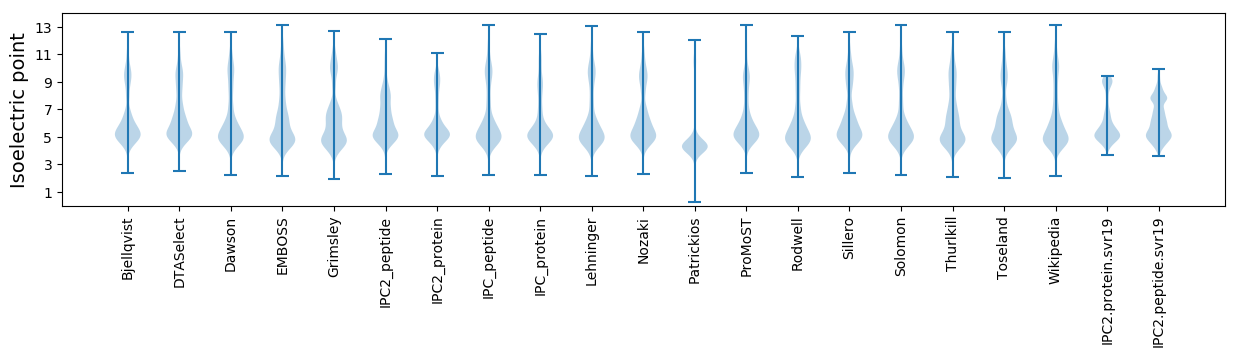
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0H4KPL3|A0A0H4KPL3_9RHOB RNA polymerase sigma factor OS=Marinovum algicola DG 898 OX=988812 GN=MALG_00285 PE=3 SV=1
MM1 pKa = 7.17QRR3 pKa = 11.84TKK5 pKa = 10.25LTLGALVASTALAGMASAQTEE26 pKa = 4.18LTLWYY31 pKa = 9.64HH32 pKa = 5.79GAGNEE37 pKa = 4.13VEE39 pKa = 4.75SNILGGIIEE48 pKa = 5.11DD49 pKa = 3.8FNGAQEE55 pKa = 4.17DD56 pKa = 3.97WQVTVEE62 pKa = 4.31SFPQEE67 pKa = 4.15SYY69 pKa = 10.96NDD71 pKa = 3.8SVVASALAGNLPCILDD87 pKa = 3.15VDD89 pKa = 4.59GPVMPNWAWAGYY101 pKa = 7.77LQPLPISQEE110 pKa = 4.25AIDD113 pKa = 3.91GHH115 pKa = 8.4LDD117 pKa = 3.28ATIGRR122 pKa = 11.84WDD124 pKa = 3.6GEE126 pKa = 4.37IYY128 pKa = 10.76SVGLWDD134 pKa = 4.73AAVALYY140 pKa = 10.38ARR142 pKa = 11.84QSTLDD147 pKa = 3.61EE148 pKa = 4.93LGLRR152 pKa = 11.84TPSLEE157 pKa = 4.19EE158 pKa = 3.68PWTKK162 pKa = 10.92EE163 pKa = 3.67EE164 pKa = 4.75FMGALEE170 pKa = 4.06AAKK173 pKa = 10.66ASGNYY178 pKa = 9.87DD179 pKa = 3.26FALDD183 pKa = 5.65LGTAWTGEE191 pKa = 3.99WYY193 pKa = 9.23PYY195 pKa = 10.83AFSPFLQSFGGDD207 pKa = 2.59IVDD210 pKa = 3.49RR211 pKa = 11.84DD212 pKa = 4.19GYY214 pKa = 8.65QTAEE218 pKa = 3.84GALNGEE224 pKa = 4.3AAMEE228 pKa = 4.63FGAWWQSLFAEE239 pKa = 6.43GYY241 pKa = 10.97APGTSQDD248 pKa = 3.48PADD251 pKa = 4.56RR252 pKa = 11.84DD253 pKa = 3.66SGFIEE258 pKa = 4.36GKK260 pKa = 10.4YY261 pKa = 9.64AFSWNGNWAAANTLASVEE279 pKa = 4.2NADD282 pKa = 4.73DD283 pKa = 4.38VVFLPAPDD291 pKa = 4.36FGNGSTIGAASWQFGVSSACEE312 pKa = 4.05HH313 pKa = 7.1PEE315 pKa = 3.61GAAAFVEE322 pKa = 4.61HH323 pKa = 7.15ALQDD327 pKa = 3.49EE328 pKa = 4.49YY329 pKa = 11.71LAAFSDD335 pKa = 5.04GIGLIPATKK344 pKa = 9.67EE345 pKa = 3.43AAQMTEE351 pKa = 4.3SYY353 pKa = 11.58AEE355 pKa = 4.32GGPLAVFFDD364 pKa = 4.56LSAEE368 pKa = 4.13QALVRR373 pKa = 11.84PVSPGYY379 pKa = 10.29VVAAKK384 pKa = 10.41VFEE387 pKa = 4.44KK388 pKa = 10.82ALSDD392 pKa = 3.46IANGADD398 pKa = 3.57VADD401 pKa = 4.21TLDD404 pKa = 3.73AAVDD408 pKa = 4.35EE409 pKa = 4.86IDD411 pKa = 5.8DD412 pKa = 5.59DD413 pKa = 4.13IEE415 pKa = 4.99ANQGYY420 pKa = 8.33GHH422 pKa = 7.53
MM1 pKa = 7.17QRR3 pKa = 11.84TKK5 pKa = 10.25LTLGALVASTALAGMASAQTEE26 pKa = 4.18LTLWYY31 pKa = 9.64HH32 pKa = 5.79GAGNEE37 pKa = 4.13VEE39 pKa = 4.75SNILGGIIEE48 pKa = 5.11DD49 pKa = 3.8FNGAQEE55 pKa = 4.17DD56 pKa = 3.97WQVTVEE62 pKa = 4.31SFPQEE67 pKa = 4.15SYY69 pKa = 10.96NDD71 pKa = 3.8SVVASALAGNLPCILDD87 pKa = 3.15VDD89 pKa = 4.59GPVMPNWAWAGYY101 pKa = 7.77LQPLPISQEE110 pKa = 4.25AIDD113 pKa = 3.91GHH115 pKa = 8.4LDD117 pKa = 3.28ATIGRR122 pKa = 11.84WDD124 pKa = 3.6GEE126 pKa = 4.37IYY128 pKa = 10.76SVGLWDD134 pKa = 4.73AAVALYY140 pKa = 10.38ARR142 pKa = 11.84QSTLDD147 pKa = 3.61EE148 pKa = 4.93LGLRR152 pKa = 11.84TPSLEE157 pKa = 4.19EE158 pKa = 3.68PWTKK162 pKa = 10.92EE163 pKa = 3.67EE164 pKa = 4.75FMGALEE170 pKa = 4.06AAKK173 pKa = 10.66ASGNYY178 pKa = 9.87DD179 pKa = 3.26FALDD183 pKa = 5.65LGTAWTGEE191 pKa = 3.99WYY193 pKa = 9.23PYY195 pKa = 10.83AFSPFLQSFGGDD207 pKa = 2.59IVDD210 pKa = 3.49RR211 pKa = 11.84DD212 pKa = 4.19GYY214 pKa = 8.65QTAEE218 pKa = 3.84GALNGEE224 pKa = 4.3AAMEE228 pKa = 4.63FGAWWQSLFAEE239 pKa = 6.43GYY241 pKa = 10.97APGTSQDD248 pKa = 3.48PADD251 pKa = 4.56RR252 pKa = 11.84DD253 pKa = 3.66SGFIEE258 pKa = 4.36GKK260 pKa = 10.4YY261 pKa = 9.64AFSWNGNWAAANTLASVEE279 pKa = 4.2NADD282 pKa = 4.73DD283 pKa = 4.38VVFLPAPDD291 pKa = 4.36FGNGSTIGAASWQFGVSSACEE312 pKa = 4.05HH313 pKa = 7.1PEE315 pKa = 3.61GAAAFVEE322 pKa = 4.61HH323 pKa = 7.15ALQDD327 pKa = 3.49EE328 pKa = 4.49YY329 pKa = 11.71LAAFSDD335 pKa = 5.04GIGLIPATKK344 pKa = 9.67EE345 pKa = 3.43AAQMTEE351 pKa = 4.3SYY353 pKa = 11.58AEE355 pKa = 4.32GGPLAVFFDD364 pKa = 4.56LSAEE368 pKa = 4.13QALVRR373 pKa = 11.84PVSPGYY379 pKa = 10.29VVAAKK384 pKa = 10.41VFEE387 pKa = 4.44KK388 pKa = 10.82ALSDD392 pKa = 3.46IANGADD398 pKa = 3.57VADD401 pKa = 4.21TLDD404 pKa = 3.73AAVDD408 pKa = 4.35EE409 pKa = 4.86IDD411 pKa = 5.8DD412 pKa = 5.59DD413 pKa = 4.13IEE415 pKa = 4.99ANQGYY420 pKa = 8.33GHH422 pKa = 7.53
Molecular weight: 44.87 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0H4KZ01|A0A0H4KZ01_9RHOB Serine phosphatase RsbU regulator of sigma subunit OS=Marinovum algicola DG 898 OX=988812 GN=MALG_03562 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1570877 |
30 |
2735 |
315.7 |
34.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.847 ± 0.055 | 0.92 ± 0.01 |
5.967 ± 0.035 | 6.042 ± 0.035 |
3.698 ± 0.021 | 8.87 ± 0.032 |
2.062 ± 0.017 | 4.957 ± 0.026 |
2.86 ± 0.027 | 10.188 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.699 ± 0.018 | 2.427 ± 0.02 |
5.2 ± 0.026 | 3.137 ± 0.016 |
7.048 ± 0.038 | 4.918 ± 0.023 |
5.353 ± 0.025 | 7.217 ± 0.033 |
1.398 ± 0.014 | 2.193 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
