
Penicillium aurantiogriseum partitivirus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; unclassified Partitiviridae
Average proteome isoelectric point is 6.87
Get precalculated fractions of proteins
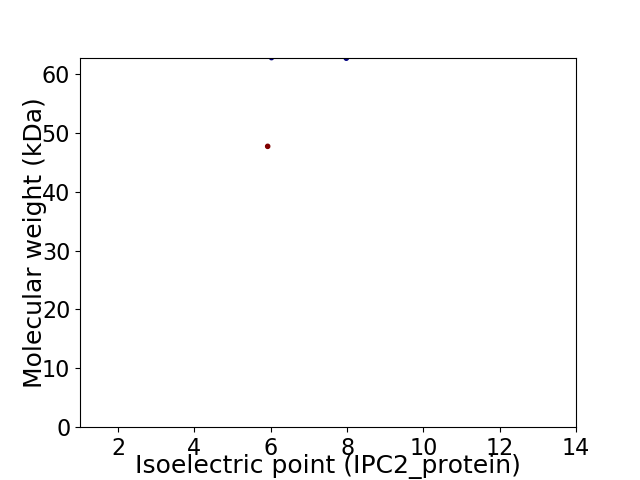
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0S2KQ39|A0A0S2KQ39_9VIRU 47 kDa protein OS=Penicillium aurantiogriseum partitivirus 1 OX=1756157 PE=4 SV=1
MM1 pKa = 7.65PRR3 pKa = 11.84QPSEE7 pKa = 4.31VPSTLAPSDD16 pKa = 3.77SASAAGSKK24 pKa = 9.95RR25 pKa = 11.84SKK27 pKa = 9.81PGKK30 pKa = 9.73AEE32 pKa = 3.64RR33 pKa = 11.84AARR36 pKa = 11.84RR37 pKa = 11.84ATGSQAPKK45 pKa = 9.75PADD48 pKa = 3.37AGKK51 pKa = 10.96AMTFASGGMIPKK63 pKa = 8.45PQPGKK68 pKa = 10.58FPVVFQTGAGEE79 pKa = 3.96PSRR82 pKa = 11.84DD83 pKa = 3.2QGFAIEE89 pKa = 4.5PNVLADD95 pKa = 3.84TVSQFPSRR103 pKa = 11.84FTSNAGYY110 pKa = 11.13AEE112 pKa = 4.25FLTFTDD118 pKa = 5.24LDD120 pKa = 4.32DD121 pKa = 6.72DD122 pKa = 5.53DD123 pKa = 4.79FAKK126 pKa = 10.37QLKK129 pKa = 9.61SASLLRR135 pKa = 11.84LAQQLVHH142 pKa = 6.26AHH144 pKa = 5.33VNMGLPQGDD153 pKa = 4.3FAPVASTEE161 pKa = 3.93VRR163 pKa = 11.84VPASVNAYY171 pKa = 9.54LSQFGEE177 pKa = 4.24FSVPALGTRR186 pKa = 11.84FLFDD190 pKa = 5.13DD191 pKa = 3.84YY192 pKa = 11.82ASTVKK197 pKa = 10.69SIIHH201 pKa = 5.37TAEE204 pKa = 3.68QLLAASNDD212 pKa = 3.4DD213 pKa = 3.66SEE215 pKa = 7.05AILKK219 pKa = 9.94RR220 pKa = 11.84CWLPMSRR227 pKa = 11.84TDD229 pKa = 2.71GHH231 pKa = 5.59TRR233 pKa = 11.84QIIANYY239 pKa = 8.93LNQWLLTNEE248 pKa = 3.73IMYY251 pKa = 8.33PAQLLEE257 pKa = 4.18KK258 pKa = 9.8AVMSGTPPAAWDD270 pKa = 3.68KK271 pKa = 10.56IKK273 pKa = 10.66PVFGDD278 pKa = 3.47TEE280 pKa = 4.37TEE282 pKa = 3.84KK283 pKa = 11.04NRR285 pKa = 11.84FDD287 pKa = 4.14FLFSGYY293 pKa = 9.57RR294 pKa = 11.84DD295 pKa = 3.47APHH298 pKa = 6.7FVTTFTTAASSAVLEE313 pKa = 4.62EE314 pKa = 5.71IGLSWDD320 pKa = 3.71NPSAAHH326 pKa = 6.71VDD328 pKa = 3.04WTFNAKK334 pKa = 9.97EE335 pKa = 4.21SFTRR339 pKa = 11.84LSDD342 pKa = 2.68NWARR346 pKa = 11.84ISTTYY351 pKa = 11.02GRR353 pKa = 11.84FFEE356 pKa = 4.63MPSSQMNRR364 pKa = 11.84SAATGSQSQMAIVSSKK380 pKa = 11.3DD381 pKa = 3.01SVTVVKK387 pKa = 9.0THH389 pKa = 6.99LALSAPEE396 pKa = 4.16FSLVACFPATCLFSGDD412 pKa = 3.52LRR414 pKa = 11.84RR415 pKa = 11.84HH416 pKa = 4.79VVVTTPLSTVQRR428 pKa = 11.84ATEE431 pKa = 4.42FVQLDD436 pKa = 3.41WRR438 pKa = 4.22
MM1 pKa = 7.65PRR3 pKa = 11.84QPSEE7 pKa = 4.31VPSTLAPSDD16 pKa = 3.77SASAAGSKK24 pKa = 9.95RR25 pKa = 11.84SKK27 pKa = 9.81PGKK30 pKa = 9.73AEE32 pKa = 3.64RR33 pKa = 11.84AARR36 pKa = 11.84RR37 pKa = 11.84ATGSQAPKK45 pKa = 9.75PADD48 pKa = 3.37AGKK51 pKa = 10.96AMTFASGGMIPKK63 pKa = 8.45PQPGKK68 pKa = 10.58FPVVFQTGAGEE79 pKa = 3.96PSRR82 pKa = 11.84DD83 pKa = 3.2QGFAIEE89 pKa = 4.5PNVLADD95 pKa = 3.84TVSQFPSRR103 pKa = 11.84FTSNAGYY110 pKa = 11.13AEE112 pKa = 4.25FLTFTDD118 pKa = 5.24LDD120 pKa = 4.32DD121 pKa = 6.72DD122 pKa = 5.53DD123 pKa = 4.79FAKK126 pKa = 10.37QLKK129 pKa = 9.61SASLLRR135 pKa = 11.84LAQQLVHH142 pKa = 6.26AHH144 pKa = 5.33VNMGLPQGDD153 pKa = 4.3FAPVASTEE161 pKa = 3.93VRR163 pKa = 11.84VPASVNAYY171 pKa = 9.54LSQFGEE177 pKa = 4.24FSVPALGTRR186 pKa = 11.84FLFDD190 pKa = 5.13DD191 pKa = 3.84YY192 pKa = 11.82ASTVKK197 pKa = 10.69SIIHH201 pKa = 5.37TAEE204 pKa = 3.68QLLAASNDD212 pKa = 3.4DD213 pKa = 3.66SEE215 pKa = 7.05AILKK219 pKa = 9.94RR220 pKa = 11.84CWLPMSRR227 pKa = 11.84TDD229 pKa = 2.71GHH231 pKa = 5.59TRR233 pKa = 11.84QIIANYY239 pKa = 8.93LNQWLLTNEE248 pKa = 3.73IMYY251 pKa = 8.33PAQLLEE257 pKa = 4.18KK258 pKa = 9.8AVMSGTPPAAWDD270 pKa = 3.68KK271 pKa = 10.56IKK273 pKa = 10.66PVFGDD278 pKa = 3.47TEE280 pKa = 4.37TEE282 pKa = 3.84KK283 pKa = 11.04NRR285 pKa = 11.84FDD287 pKa = 4.14FLFSGYY293 pKa = 9.57RR294 pKa = 11.84DD295 pKa = 3.47APHH298 pKa = 6.7FVTTFTTAASSAVLEE313 pKa = 4.62EE314 pKa = 5.71IGLSWDD320 pKa = 3.71NPSAAHH326 pKa = 6.71VDD328 pKa = 3.04WTFNAKK334 pKa = 9.97EE335 pKa = 4.21SFTRR339 pKa = 11.84LSDD342 pKa = 2.68NWARR346 pKa = 11.84ISTTYY351 pKa = 11.02GRR353 pKa = 11.84FFEE356 pKa = 4.63MPSSQMNRR364 pKa = 11.84SAATGSQSQMAIVSSKK380 pKa = 11.3DD381 pKa = 3.01SVTVVKK387 pKa = 9.0THH389 pKa = 6.99LALSAPEE396 pKa = 4.16FSLVACFPATCLFSGDD412 pKa = 3.52LRR414 pKa = 11.84RR415 pKa = 11.84HH416 pKa = 4.79VVVTTPLSTVQRR428 pKa = 11.84ATEE431 pKa = 4.42FVQLDD436 pKa = 3.41WRR438 pKa = 4.22
Molecular weight: 47.7 kDa
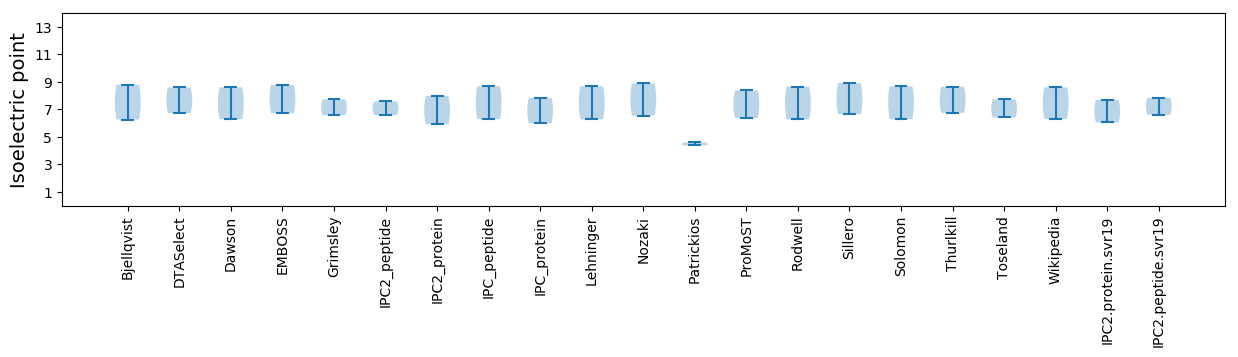
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0S2KQ39|A0A0S2KQ39_9VIRU 47 kDa protein OS=Penicillium aurantiogriseum partitivirus 1 OX=1756157 PE=4 SV=1
MM1 pKa = 7.9AFTQEE6 pKa = 4.1PTQHH10 pKa = 5.69YY11 pKa = 9.06VLAKK15 pKa = 9.8GSHH18 pKa = 6.8LIDD21 pKa = 3.96SLHH24 pKa = 6.47LRR26 pKa = 11.84PAKK29 pKa = 10.42AGSATSEE36 pKa = 4.2DD37 pKa = 4.2VLPTGYY43 pKa = 10.64NSPNLRR49 pKa = 11.84EE50 pKa = 3.64IAKK53 pKa = 10.08FGGYY57 pKa = 7.52STYY60 pKa = 11.18SSASNTDD67 pKa = 2.61PWVRR71 pKa = 11.84EE72 pKa = 3.87TLKK75 pKa = 10.74LFSRR79 pKa = 11.84EE80 pKa = 3.86RR81 pKa = 11.84YY82 pKa = 8.44EE83 pKa = 4.06EE84 pKa = 3.99IYY86 pKa = 10.92GFTRR90 pKa = 11.84RR91 pKa = 11.84PEE93 pKa = 4.26GTPGMYY99 pKa = 10.1KK100 pKa = 10.27SLAKK104 pKa = 10.14FAGEE108 pKa = 3.83KK109 pKa = 9.93SHH111 pKa = 7.31FRR113 pKa = 11.84DD114 pKa = 3.75LTVSQQKK121 pKa = 10.56AMRR124 pKa = 11.84RR125 pKa = 11.84SIAKK129 pKa = 9.5AKK131 pKa = 10.33KK132 pKa = 9.94AFKK135 pKa = 10.34LPYY138 pKa = 9.55KK139 pKa = 10.34RR140 pKa = 11.84EE141 pKa = 3.91PLDD144 pKa = 3.15WHH146 pKa = 6.48EE147 pKa = 4.21VGQFLRR153 pKa = 11.84RR154 pKa = 11.84DD155 pKa = 3.54TSAGSTFMGQKK166 pKa = 10.07KK167 pKa = 9.93GDD169 pKa = 3.65VMEE172 pKa = 5.9DD173 pKa = 2.98IYY175 pKa = 11.47HH176 pKa = 6.14EE177 pKa = 4.38ARR179 pKa = 11.84WLGHH183 pKa = 6.06RR184 pKa = 11.84MKK186 pKa = 10.62QDD188 pKa = 3.31GEE190 pKa = 4.54SSFNPTKK197 pKa = 10.28MRR199 pKa = 11.84FPPCLAGQRR208 pKa = 11.84GGMSEE213 pKa = 4.57RR214 pKa = 11.84DD215 pKa = 3.57DD216 pKa = 3.99PKK218 pKa = 11.09TRR220 pKa = 11.84LVWIYY225 pKa = 8.98PAEE228 pKa = 3.96MLVIEE233 pKa = 5.06GFYY236 pKa = 10.8APLMYY241 pKa = 9.7RR242 pKa = 11.84DD243 pKa = 4.6FMNDD247 pKa = 2.96PNSPMLNGKK256 pKa = 8.05SAPRR260 pKa = 11.84LYY262 pKa = 11.01AEE264 pKa = 4.49WCCGLRR270 pKa = 11.84EE271 pKa = 4.19GEE273 pKa = 4.23TLYY276 pKa = 11.53GLDD279 pKa = 4.6FSAFDD284 pKa = 3.67TKK286 pKa = 11.08VPTWLIYY293 pKa = 9.48TAFDD297 pKa = 3.36ILRR300 pKa = 11.84QNIEE304 pKa = 3.32WSTFQGKK311 pKa = 8.11PVSKK315 pKa = 10.53QDD317 pKa = 3.2AQKK320 pKa = 9.9WRR322 pKa = 11.84NVWDD326 pKa = 3.59GMVWYY331 pKa = 9.07FVNTPILMPDD341 pKa = 2.41GRR343 pKa = 11.84MFRR346 pKa = 11.84KK347 pKa = 9.98YY348 pKa = 10.14RR349 pKa = 11.84GVPSGSWWTQMIDD362 pKa = 3.49SVVNYY367 pKa = 10.26ILIDD371 pKa = 3.69YY372 pKa = 9.7LAEE375 pKa = 4.23CQEE378 pKa = 4.02VEE380 pKa = 3.92IRR382 pKa = 11.84NLRR385 pKa = 11.84VLGDD389 pKa = 3.68DD390 pKa = 3.44SAFRR394 pKa = 11.84STDD397 pKa = 3.29QFSLEE402 pKa = 4.02QAKK405 pKa = 10.08VDD407 pKa = 4.07CEE409 pKa = 4.19PTHH412 pKa = 6.37MLLKK416 pKa = 10.35PEE418 pKa = 4.25KK419 pKa = 10.11CEE421 pKa = 3.96KK422 pKa = 10.32TKK424 pKa = 10.91DD425 pKa = 3.32PCEE428 pKa = 4.01FKK430 pKa = 11.23LLGTTYY436 pKa = 10.79RR437 pKa = 11.84DD438 pKa = 3.59GRR440 pKa = 11.84VHH442 pKa = 7.37RR443 pKa = 11.84PTEE446 pKa = 3.75EE447 pKa = 3.53WFKK450 pKa = 11.17LVIYY454 pKa = 9.67PEE456 pKa = 4.57SSVHH460 pKa = 5.71TLDD463 pKa = 3.85ISFTRR468 pKa = 11.84LIGLWLGGAMWDD480 pKa = 4.48KK481 pKa = 10.62EE482 pKa = 4.18FCRR485 pKa = 11.84YY486 pKa = 8.64MDD488 pKa = 4.7FFQSSYY494 pKa = 10.42PCPEE498 pKa = 4.29EE499 pKa = 4.24GWFSKK504 pKa = 10.13DD505 pKa = 3.02QKK507 pKa = 10.68RR508 pKa = 11.84WLEE511 pKa = 3.93VIYY514 pKa = 10.54SGKK517 pKa = 10.27APRR520 pKa = 11.84GWTTKK525 pKa = 10.44RR526 pKa = 11.84SLFWRR531 pKa = 11.84SIFYY535 pKa = 10.51AYY537 pKa = 10.44GG538 pKa = 3.3
MM1 pKa = 7.9AFTQEE6 pKa = 4.1PTQHH10 pKa = 5.69YY11 pKa = 9.06VLAKK15 pKa = 9.8GSHH18 pKa = 6.8LIDD21 pKa = 3.96SLHH24 pKa = 6.47LRR26 pKa = 11.84PAKK29 pKa = 10.42AGSATSEE36 pKa = 4.2DD37 pKa = 4.2VLPTGYY43 pKa = 10.64NSPNLRR49 pKa = 11.84EE50 pKa = 3.64IAKK53 pKa = 10.08FGGYY57 pKa = 7.52STYY60 pKa = 11.18SSASNTDD67 pKa = 2.61PWVRR71 pKa = 11.84EE72 pKa = 3.87TLKK75 pKa = 10.74LFSRR79 pKa = 11.84EE80 pKa = 3.86RR81 pKa = 11.84YY82 pKa = 8.44EE83 pKa = 4.06EE84 pKa = 3.99IYY86 pKa = 10.92GFTRR90 pKa = 11.84RR91 pKa = 11.84PEE93 pKa = 4.26GTPGMYY99 pKa = 10.1KK100 pKa = 10.27SLAKK104 pKa = 10.14FAGEE108 pKa = 3.83KK109 pKa = 9.93SHH111 pKa = 7.31FRR113 pKa = 11.84DD114 pKa = 3.75LTVSQQKK121 pKa = 10.56AMRR124 pKa = 11.84RR125 pKa = 11.84SIAKK129 pKa = 9.5AKK131 pKa = 10.33KK132 pKa = 9.94AFKK135 pKa = 10.34LPYY138 pKa = 9.55KK139 pKa = 10.34RR140 pKa = 11.84EE141 pKa = 3.91PLDD144 pKa = 3.15WHH146 pKa = 6.48EE147 pKa = 4.21VGQFLRR153 pKa = 11.84RR154 pKa = 11.84DD155 pKa = 3.54TSAGSTFMGQKK166 pKa = 10.07KK167 pKa = 9.93GDD169 pKa = 3.65VMEE172 pKa = 5.9DD173 pKa = 2.98IYY175 pKa = 11.47HH176 pKa = 6.14EE177 pKa = 4.38ARR179 pKa = 11.84WLGHH183 pKa = 6.06RR184 pKa = 11.84MKK186 pKa = 10.62QDD188 pKa = 3.31GEE190 pKa = 4.54SSFNPTKK197 pKa = 10.28MRR199 pKa = 11.84FPPCLAGQRR208 pKa = 11.84GGMSEE213 pKa = 4.57RR214 pKa = 11.84DD215 pKa = 3.57DD216 pKa = 3.99PKK218 pKa = 11.09TRR220 pKa = 11.84LVWIYY225 pKa = 8.98PAEE228 pKa = 3.96MLVIEE233 pKa = 5.06GFYY236 pKa = 10.8APLMYY241 pKa = 9.7RR242 pKa = 11.84DD243 pKa = 4.6FMNDD247 pKa = 2.96PNSPMLNGKK256 pKa = 8.05SAPRR260 pKa = 11.84LYY262 pKa = 11.01AEE264 pKa = 4.49WCCGLRR270 pKa = 11.84EE271 pKa = 4.19GEE273 pKa = 4.23TLYY276 pKa = 11.53GLDD279 pKa = 4.6FSAFDD284 pKa = 3.67TKK286 pKa = 11.08VPTWLIYY293 pKa = 9.48TAFDD297 pKa = 3.36ILRR300 pKa = 11.84QNIEE304 pKa = 3.32WSTFQGKK311 pKa = 8.11PVSKK315 pKa = 10.53QDD317 pKa = 3.2AQKK320 pKa = 9.9WRR322 pKa = 11.84NVWDD326 pKa = 3.59GMVWYY331 pKa = 9.07FVNTPILMPDD341 pKa = 2.41GRR343 pKa = 11.84MFRR346 pKa = 11.84KK347 pKa = 9.98YY348 pKa = 10.14RR349 pKa = 11.84GVPSGSWWTQMIDD362 pKa = 3.49SVVNYY367 pKa = 10.26ILIDD371 pKa = 3.69YY372 pKa = 9.7LAEE375 pKa = 4.23CQEE378 pKa = 4.02VEE380 pKa = 3.92IRR382 pKa = 11.84NLRR385 pKa = 11.84VLGDD389 pKa = 3.68DD390 pKa = 3.44SAFRR394 pKa = 11.84STDD397 pKa = 3.29QFSLEE402 pKa = 4.02QAKK405 pKa = 10.08VDD407 pKa = 4.07CEE409 pKa = 4.19PTHH412 pKa = 6.37MLLKK416 pKa = 10.35PEE418 pKa = 4.25KK419 pKa = 10.11CEE421 pKa = 3.96KK422 pKa = 10.32TKK424 pKa = 10.91DD425 pKa = 3.32PCEE428 pKa = 4.01FKK430 pKa = 11.23LLGTTYY436 pKa = 10.79RR437 pKa = 11.84DD438 pKa = 3.59GRR440 pKa = 11.84VHH442 pKa = 7.37RR443 pKa = 11.84PTEE446 pKa = 3.75EE447 pKa = 3.53WFKK450 pKa = 11.17LVIYY454 pKa = 9.67PEE456 pKa = 4.57SSVHH460 pKa = 5.71TLDD463 pKa = 3.85ISFTRR468 pKa = 11.84LIGLWLGGAMWDD480 pKa = 4.48KK481 pKa = 10.62EE482 pKa = 4.18FCRR485 pKa = 11.84YY486 pKa = 8.64MDD488 pKa = 4.7FFQSSYY494 pKa = 10.42PCPEE498 pKa = 4.29EE499 pKa = 4.24GWFSKK504 pKa = 10.13DD505 pKa = 3.02QKK507 pKa = 10.68RR508 pKa = 11.84WLEE511 pKa = 3.93VIYY514 pKa = 10.54SGKK517 pKa = 10.27APRR520 pKa = 11.84GWTTKK525 pKa = 10.44RR526 pKa = 11.84SLFWRR531 pKa = 11.84SIFYY535 pKa = 10.51AYY537 pKa = 10.44GG538 pKa = 3.3
Molecular weight: 62.67 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
976 |
438 |
538 |
488.0 |
55.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.402 ± 2.425 | 1.23 ± 0.357 |
5.738 ± 0.02 | 5.635 ± 0.851 |
5.943 ± 0.295 | 6.25 ± 0.655 |
1.844 ± 0.012 | 3.279 ± 0.353 |
5.533 ± 0.933 | 7.684 ± 0.098 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.971 ± 0.451 | 2.561 ± 0.267 |
6.148 ± 0.31 | 3.893 ± 0.441 |
6.352 ± 0.722 | 8.607 ± 1.093 |
6.66 ± 0.723 | 5.225 ± 0.765 |
2.664 ± 0.699 | 3.381 ± 1.169 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
