
Nigrospora oryzae victorivirus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Totiviridae; Victorivirus; unclassified Victorivirus
Average proteome isoelectric point is 6.67
Get precalculated fractions of proteins
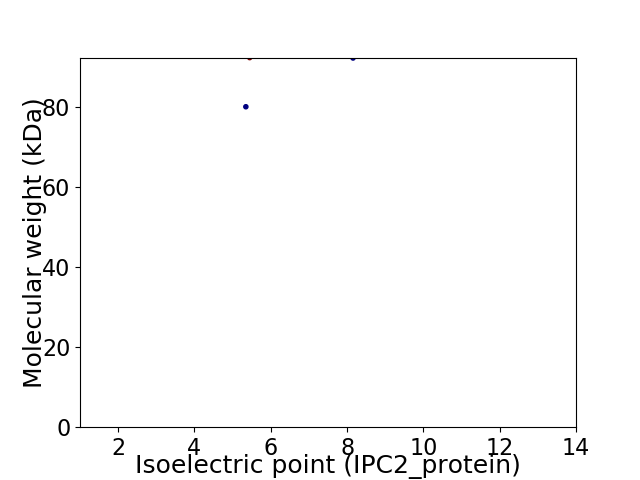
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0S3IRQ6|A0A0S3IRQ6_9VIRU RNA-directed RNA polymerase OS=Nigrospora oryzae victorivirus 1 OX=1765736 GN=RdRp PE=3 SV=1
MM1 pKa = 7.73ASTANFQVSVTSLLTGAVSGTSGGLIQTDD30 pKa = 3.72GTFRR34 pKa = 11.84RR35 pKa = 11.84YY36 pKa = 9.89RR37 pKa = 11.84SGLSIGVQEE46 pKa = 4.7HH47 pKa = 6.07GVLNYY52 pKa = 9.79SRR54 pKa = 11.84RR55 pKa = 11.84SIFYY59 pKa = 9.59EE60 pKa = 3.49VGRR63 pKa = 11.84RR64 pKa = 11.84YY65 pKa = 10.5GRR67 pKa = 11.84LTDD70 pKa = 3.55ALAYY74 pKa = 10.33QKK76 pKa = 11.19GDD78 pKa = 3.79DD79 pKa = 4.32EE80 pKa = 4.79GLPLIDD86 pKa = 4.55ASVPINPAQAANFEE100 pKa = 4.05GWARR104 pKa = 11.84KK105 pKa = 9.44YY106 pKa = 11.34SNFSPQWATMDD117 pKa = 3.85LAGLAEE123 pKa = 4.2RR124 pKa = 11.84LAKK127 pKa = 10.19GVAAQSVYY135 pKa = 11.25GGVTTTNLRR144 pKa = 11.84GGQPVRR150 pKa = 11.84VVALGTLDD158 pKa = 3.82SPQTASTNSIFIPRR172 pKa = 11.84TVDD175 pKa = 2.78TTTSDD180 pKa = 2.93NTFAVLAAAANGEE193 pKa = 4.38GATVTTDD200 pKa = 3.42VVRR203 pKa = 11.84LDD205 pKa = 3.42AATNQPVIPAVDD217 pKa = 4.79GYY219 pKa = 11.65AFATACVEE227 pKa = 3.8ALRR230 pKa = 11.84IVGANLEE237 pKa = 4.16EE238 pKa = 4.95SGAGDD243 pKa = 3.32VFAYY247 pKa = 10.22AVARR251 pKa = 11.84GIHH254 pKa = 6.41AIVSVVAHH262 pKa = 6.09TDD264 pKa = 2.83EE265 pKa = 4.95GGWMRR270 pKa = 11.84NAFRR274 pKa = 11.84SCTFRR279 pKa = 11.84VPYY282 pKa = 10.55GGINSALSQYY292 pKa = 9.81PQLPPLAGSSAASVSSWVDD311 pKa = 3.28AVALKK316 pKa = 9.07TAAAVAHH323 pKa = 6.88CDD325 pKa = 3.35PCVTATGGLYY335 pKa = 7.99PTVFTSSSGQVGAAGTAGDD354 pKa = 4.82DD355 pKa = 3.79PTNADD360 pKa = 3.52AQDD363 pKa = 3.5VARR366 pKa = 11.84QLAADD371 pKa = 4.88FGRR374 pKa = 11.84FAPTYY379 pKa = 7.99ITALRR384 pKa = 11.84RR385 pKa = 11.84IFGLTSDD392 pKa = 3.89SGVAAMCFSTAARR405 pKa = 11.84QALFGPPDD413 pKa = 3.04RR414 pKa = 11.84HH415 pKa = 6.24LKK417 pKa = 9.46YY418 pKa = 9.72KK419 pKa = 9.89TVAPYY424 pKa = 9.78FWIEE428 pKa = 3.9PTSLIPASAFGTTAEE443 pKa = 4.3SAGFGALTTAGIEE456 pKa = 4.03TTIPCFEE463 pKa = 4.28RR464 pKa = 11.84VRR466 pKa = 11.84EE467 pKa = 3.99LDD469 pKa = 2.82HH470 pKa = 6.78GRR472 pKa = 11.84NANFSTIAFKK482 pKa = 10.48MRR484 pKa = 11.84TARR487 pKa = 11.84TSGLVAAYY495 pKa = 9.86AGAPAALAGLKK506 pKa = 10.08LYY508 pKa = 10.74QFDD511 pKa = 4.04EE512 pKa = 5.08DD513 pKa = 4.35SVALAGDD520 pKa = 3.59QGPTPGDD527 pKa = 3.14VPMKK531 pKa = 10.41HH532 pKa = 6.08SQADD536 pKa = 3.72PLSSYY541 pKa = 10.7LWTRR545 pKa = 11.84GQSPIPAPAEE555 pKa = 4.02FINIQGAYY563 pKa = 7.45AAKK566 pKa = 10.26YY567 pKa = 10.18KK568 pKa = 10.5IVDD571 pKa = 3.34WDD573 pKa = 3.71DD574 pKa = 3.81DD575 pKa = 4.54FNGRR579 pKa = 11.84LGDD582 pKa = 3.68LPEE585 pKa = 4.69AFEE588 pKa = 4.53LEE590 pKa = 4.32NYY592 pKa = 9.27GIRR595 pKa = 11.84WRR597 pKa = 11.84VTVPTALPSGASNAGDD613 pKa = 3.4TGAKK617 pKa = 9.24RR618 pKa = 11.84ARR620 pKa = 11.84TRR622 pKa = 11.84AAIALAQANIRR633 pKa = 11.84NRR635 pKa = 11.84GFGEE639 pKa = 4.07ANSPVISVSNVPPSFDD655 pKa = 3.54DD656 pKa = 3.7TPPPMMAAASGAVHH670 pKa = 7.23DD671 pKa = 4.72AANRR675 pKa = 11.84TIPEE679 pKa = 4.2YY680 pKa = 11.32AEE682 pKa = 4.25GAATAGAPPVMGAPLGPIPHH702 pKa = 6.41HH703 pKa = 5.9QPLRR707 pKa = 11.84GAPYY711 pKa = 8.7PRR713 pKa = 11.84QGGGQVGGAGLPPNPPLPGNNPPPPPPTGPSNPPTNPDD751 pKa = 3.51LDD753 pKa = 4.21YY754 pKa = 11.33QPDD757 pKa = 3.82PAGPEE762 pKa = 3.81AAHH765 pKa = 6.64APPAGQVV772 pKa = 2.93
MM1 pKa = 7.73ASTANFQVSVTSLLTGAVSGTSGGLIQTDD30 pKa = 3.72GTFRR34 pKa = 11.84RR35 pKa = 11.84YY36 pKa = 9.89RR37 pKa = 11.84SGLSIGVQEE46 pKa = 4.7HH47 pKa = 6.07GVLNYY52 pKa = 9.79SRR54 pKa = 11.84RR55 pKa = 11.84SIFYY59 pKa = 9.59EE60 pKa = 3.49VGRR63 pKa = 11.84RR64 pKa = 11.84YY65 pKa = 10.5GRR67 pKa = 11.84LTDD70 pKa = 3.55ALAYY74 pKa = 10.33QKK76 pKa = 11.19GDD78 pKa = 3.79DD79 pKa = 4.32EE80 pKa = 4.79GLPLIDD86 pKa = 4.55ASVPINPAQAANFEE100 pKa = 4.05GWARR104 pKa = 11.84KK105 pKa = 9.44YY106 pKa = 11.34SNFSPQWATMDD117 pKa = 3.85LAGLAEE123 pKa = 4.2RR124 pKa = 11.84LAKK127 pKa = 10.19GVAAQSVYY135 pKa = 11.25GGVTTTNLRR144 pKa = 11.84GGQPVRR150 pKa = 11.84VVALGTLDD158 pKa = 3.82SPQTASTNSIFIPRR172 pKa = 11.84TVDD175 pKa = 2.78TTTSDD180 pKa = 2.93NTFAVLAAAANGEE193 pKa = 4.38GATVTTDD200 pKa = 3.42VVRR203 pKa = 11.84LDD205 pKa = 3.42AATNQPVIPAVDD217 pKa = 4.79GYY219 pKa = 11.65AFATACVEE227 pKa = 3.8ALRR230 pKa = 11.84IVGANLEE237 pKa = 4.16EE238 pKa = 4.95SGAGDD243 pKa = 3.32VFAYY247 pKa = 10.22AVARR251 pKa = 11.84GIHH254 pKa = 6.41AIVSVVAHH262 pKa = 6.09TDD264 pKa = 2.83EE265 pKa = 4.95GGWMRR270 pKa = 11.84NAFRR274 pKa = 11.84SCTFRR279 pKa = 11.84VPYY282 pKa = 10.55GGINSALSQYY292 pKa = 9.81PQLPPLAGSSAASVSSWVDD311 pKa = 3.28AVALKK316 pKa = 9.07TAAAVAHH323 pKa = 6.88CDD325 pKa = 3.35PCVTATGGLYY335 pKa = 7.99PTVFTSSSGQVGAAGTAGDD354 pKa = 4.82DD355 pKa = 3.79PTNADD360 pKa = 3.52AQDD363 pKa = 3.5VARR366 pKa = 11.84QLAADD371 pKa = 4.88FGRR374 pKa = 11.84FAPTYY379 pKa = 7.99ITALRR384 pKa = 11.84RR385 pKa = 11.84IFGLTSDD392 pKa = 3.89SGVAAMCFSTAARR405 pKa = 11.84QALFGPPDD413 pKa = 3.04RR414 pKa = 11.84HH415 pKa = 6.24LKK417 pKa = 9.46YY418 pKa = 9.72KK419 pKa = 9.89TVAPYY424 pKa = 9.78FWIEE428 pKa = 3.9PTSLIPASAFGTTAEE443 pKa = 4.3SAGFGALTTAGIEE456 pKa = 4.03TTIPCFEE463 pKa = 4.28RR464 pKa = 11.84VRR466 pKa = 11.84EE467 pKa = 3.99LDD469 pKa = 2.82HH470 pKa = 6.78GRR472 pKa = 11.84NANFSTIAFKK482 pKa = 10.48MRR484 pKa = 11.84TARR487 pKa = 11.84TSGLVAAYY495 pKa = 9.86AGAPAALAGLKK506 pKa = 10.08LYY508 pKa = 10.74QFDD511 pKa = 4.04EE512 pKa = 5.08DD513 pKa = 4.35SVALAGDD520 pKa = 3.59QGPTPGDD527 pKa = 3.14VPMKK531 pKa = 10.41HH532 pKa = 6.08SQADD536 pKa = 3.72PLSSYY541 pKa = 10.7LWTRR545 pKa = 11.84GQSPIPAPAEE555 pKa = 4.02FINIQGAYY563 pKa = 7.45AAKK566 pKa = 10.26YY567 pKa = 10.18KK568 pKa = 10.5IVDD571 pKa = 3.34WDD573 pKa = 3.71DD574 pKa = 3.81DD575 pKa = 4.54FNGRR579 pKa = 11.84LGDD582 pKa = 3.68LPEE585 pKa = 4.69AFEE588 pKa = 4.53LEE590 pKa = 4.32NYY592 pKa = 9.27GIRR595 pKa = 11.84WRR597 pKa = 11.84VTVPTALPSGASNAGDD613 pKa = 3.4TGAKK617 pKa = 9.24RR618 pKa = 11.84ARR620 pKa = 11.84TRR622 pKa = 11.84AAIALAQANIRR633 pKa = 11.84NRR635 pKa = 11.84GFGEE639 pKa = 4.07ANSPVISVSNVPPSFDD655 pKa = 3.54DD656 pKa = 3.7TPPPMMAAASGAVHH670 pKa = 7.23DD671 pKa = 4.72AANRR675 pKa = 11.84TIPEE679 pKa = 4.2YY680 pKa = 11.32AEE682 pKa = 4.25GAATAGAPPVMGAPLGPIPHH702 pKa = 6.41HH703 pKa = 5.9QPLRR707 pKa = 11.84GAPYY711 pKa = 8.7PRR713 pKa = 11.84QGGGQVGGAGLPPNPPLPGNNPPPPPPTGPSNPPTNPDD751 pKa = 3.51LDD753 pKa = 4.21YY754 pKa = 11.33QPDD757 pKa = 3.82PAGPEE762 pKa = 3.81AAHH765 pKa = 6.64APPAGQVV772 pKa = 2.93
Molecular weight: 79.99 kDa
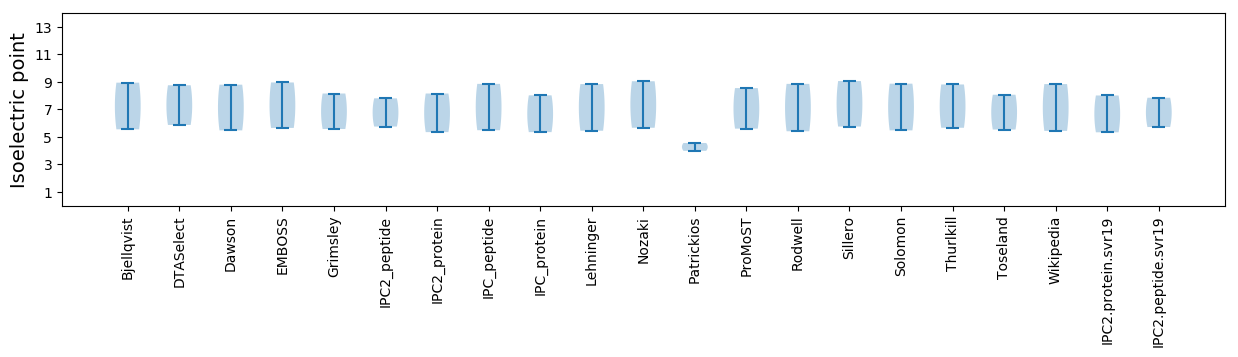
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0S3IRQ6|A0A0S3IRQ6_9VIRU RNA-directed RNA polymerase OS=Nigrospora oryzae victorivirus 1 OX=1765736 GN=RdRp PE=3 SV=1
MM1 pKa = 7.2GQRR4 pKa = 11.84ATEE7 pKa = 4.2RR8 pKa = 11.84GNDD11 pKa = 3.82PPSLGQALEE20 pKa = 4.16RR21 pKa = 11.84VARR24 pKa = 11.84RR25 pKa = 11.84MGITAKK31 pKa = 10.64LDD33 pKa = 3.3MPFADD38 pKa = 4.16QISFIYY44 pKa = 9.76RR45 pKa = 11.84PQWDD49 pKa = 3.9GRR51 pKa = 11.84PVPRR55 pKa = 11.84LARR58 pKa = 11.84AAVSYY63 pKa = 8.39LYY65 pKa = 10.6CDD67 pKa = 3.6VPVQVPLEE75 pKa = 4.3KK76 pKa = 10.51DD77 pKa = 3.04DD78 pKa = 3.78LTRR81 pKa = 11.84LLGYY85 pKa = 10.18LCEE88 pKa = 4.59HH89 pKa = 6.8LTMPPSLDD97 pKa = 3.23RR98 pKa = 11.84QTWVTDD104 pKa = 3.34KK105 pKa = 10.63KK106 pKa = 10.74RR107 pKa = 11.84AMEE110 pKa = 4.46AFPPKK115 pKa = 9.44RR116 pKa = 11.84RR117 pKa = 11.84AMALTKK123 pKa = 10.86ANIFLDD129 pKa = 4.14EE130 pKa = 4.18VARR133 pKa = 11.84DD134 pKa = 3.75LDD136 pKa = 3.24KK137 pKa = 11.55HH138 pKa = 5.23MPARR142 pKa = 11.84LLALLPYY149 pKa = 9.89LGKK152 pKa = 10.54LRR154 pKa = 11.84GVGATHH160 pKa = 7.06DD161 pKa = 3.52QATAFLLYY169 pKa = 10.65SSLLSDD175 pKa = 4.34HH176 pKa = 6.96CPLAAEE182 pKa = 4.28WGLFALTQPKK192 pKa = 8.85EE193 pKa = 4.08AKK195 pKa = 9.59EE196 pKa = 3.81VSNFLKK202 pKa = 10.81AVGANASSYY211 pKa = 10.86GALLVEE217 pKa = 4.54TDD219 pKa = 3.55TLLGRR224 pKa = 11.84DD225 pKa = 3.72TPGTDD230 pKa = 3.07LVADD234 pKa = 3.56SRR236 pKa = 11.84RR237 pKa = 11.84RR238 pKa = 11.84CDD240 pKa = 3.54LEE242 pKa = 3.89LARR245 pKa = 11.84EE246 pKa = 4.0EE247 pKa = 4.16VLAEE251 pKa = 4.17FDD253 pKa = 4.71DD254 pKa = 4.11SALRR258 pKa = 11.84SAVRR262 pKa = 11.84RR263 pKa = 11.84VLRR266 pKa = 11.84HH267 pKa = 5.33EE268 pKa = 4.29LRR270 pKa = 11.84SEE272 pKa = 3.82HH273 pKa = 5.41GSYY276 pKa = 10.31RR277 pKa = 11.84IEE279 pKa = 4.28YY280 pKa = 7.02PTQEE284 pKa = 4.28DD285 pKa = 3.6HH286 pKa = 5.84WTSRR290 pKa = 11.84WSWAVNGSHH299 pKa = 6.6SALLAGEE306 pKa = 4.25FPVPNEE312 pKa = 4.04VKK314 pKa = 10.15RR315 pKa = 11.84QVHH318 pKa = 5.55KK319 pKa = 9.87LHH321 pKa = 6.35RR322 pKa = 11.84RR323 pKa = 11.84AWLEE327 pKa = 3.89TIEE330 pKa = 4.46RR331 pKa = 11.84NPCIGWDD338 pKa = 3.02GKK340 pKa = 8.75TFVSCAPKK348 pKa = 10.27PEE350 pKa = 4.39CGKK353 pKa = 9.92IRR355 pKa = 11.84AIYY358 pKa = 10.33ACDD361 pKa = 3.68TINYY365 pKa = 8.97LAFEE369 pKa = 4.3HH370 pKa = 6.71LMAGVEE376 pKa = 4.24SRR378 pKa = 11.84WLGKK382 pKa = 10.09RR383 pKa = 11.84VILNPGRR390 pKa = 11.84GGHH393 pKa = 6.5LGIAEE398 pKa = 3.99RR399 pKa = 11.84VAACRR404 pKa = 11.84SRR406 pKa = 11.84SGISAMLDD414 pKa = 3.2YY415 pKa = 11.48DD416 pKa = 4.67DD417 pKa = 5.98FNSHH421 pKa = 5.95HH422 pKa = 5.91TTKK425 pKa = 10.93AMQILIEE432 pKa = 4.11EE433 pKa = 4.4TAALTGYY440 pKa = 9.61PPEE443 pKa = 4.15LTKK446 pKa = 10.66PLLDD450 pKa = 3.66SFEE453 pKa = 5.25KK454 pKa = 10.67EE455 pKa = 4.92DD456 pKa = 3.78IYY458 pKa = 11.45LDD460 pKa = 3.68GKK462 pKa = 10.81LVGRR466 pKa = 11.84VKK468 pKa = 9.49GTLMSGHH475 pKa = 6.99RR476 pKa = 11.84CTTYY480 pKa = 10.84VNSVLNMAYY489 pKa = 10.81LMVVLGEE496 pKa = 4.07DD497 pKa = 2.9WVMEE501 pKa = 4.17RR502 pKa = 11.84QSLHH506 pKa = 7.11VGDD509 pKa = 4.61DD510 pKa = 3.37VYY512 pKa = 11.02MGLKK516 pKa = 10.45NYY518 pKa = 10.77GEE520 pKa = 4.51TGWMISKK527 pKa = 10.28IMGSRR532 pKa = 11.84LRR534 pKa = 11.84MNRR537 pKa = 11.84RR538 pKa = 11.84KK539 pKa = 10.03QSVGHH544 pKa = 6.03VSTEE548 pKa = 3.89FLRR551 pKa = 11.84VASDD555 pKa = 3.18ARR557 pKa = 11.84DD558 pKa = 3.38SYY560 pKa = 11.92GYY562 pKa = 10.14LARR565 pKa = 11.84AAANLIAGNWYY576 pKa = 10.11SDD578 pKa = 3.18KK579 pKa = 10.97LLNPSEE585 pKa = 4.21GLITMIQGARR595 pKa = 11.84TLANRR600 pKa = 11.84ARR602 pKa = 11.84NGNAPLLLEE611 pKa = 4.3SAVKK615 pKa = 10.17RR616 pKa = 11.84VLGADD621 pKa = 3.89CPDD624 pKa = 4.23DD625 pKa = 3.39ATLRR629 pKa = 11.84HH630 pKa = 6.14MLTGSAAVNDD640 pKa = 4.53GPQYY644 pKa = 10.53LSSGRR649 pKa = 11.84HH650 pKa = 3.35RR651 pKa = 11.84RR652 pKa = 11.84VRR654 pKa = 11.84TSVVFEE660 pKa = 4.24VVDD663 pKa = 3.58TTGYY667 pKa = 8.73ATLPDD672 pKa = 4.34LASKK676 pKa = 10.78AYY678 pKa = 9.16LAKK681 pKa = 10.18HH682 pKa = 6.13ASPLEE687 pKa = 3.99IATLTSSGIDD697 pKa = 3.39PLPTMRR703 pKa = 11.84KK704 pKa = 9.3SSWTKK709 pKa = 10.73SLALSDD715 pKa = 4.33RR716 pKa = 11.84KK717 pKa = 10.61FEE719 pKa = 4.47TIVFKK724 pKa = 10.97PMEE727 pKa = 4.0QWPAVGSVQAEE738 pKa = 3.84HH739 pKa = 7.32LLRR742 pKa = 11.84EE743 pKa = 4.39RR744 pKa = 11.84KK745 pKa = 8.56PHH747 pKa = 6.0GVLLKK752 pKa = 10.9YY753 pKa = 10.14PLLVLMRR760 pKa = 11.84DD761 pKa = 4.47RR762 pKa = 11.84IPEE765 pKa = 3.91PVLRR769 pKa = 11.84VAVAAAGGRR778 pKa = 11.84DD779 pKa = 3.75YY780 pKa = 11.76VDD782 pKa = 5.79DD783 pKa = 4.54ISLEE787 pKa = 3.89AWGDD791 pKa = 3.57YY792 pKa = 11.05SNPTIVNTVLSYY804 pKa = 11.58SDD806 pKa = 4.55ASMLSKK812 pKa = 10.39RR813 pKa = 11.84ASVSVLTSRR822 pKa = 11.84RR823 pKa = 11.84RR824 pKa = 11.84CYY826 pKa = 10.4VV827 pKa = 2.75
MM1 pKa = 7.2GQRR4 pKa = 11.84ATEE7 pKa = 4.2RR8 pKa = 11.84GNDD11 pKa = 3.82PPSLGQALEE20 pKa = 4.16RR21 pKa = 11.84VARR24 pKa = 11.84RR25 pKa = 11.84MGITAKK31 pKa = 10.64LDD33 pKa = 3.3MPFADD38 pKa = 4.16QISFIYY44 pKa = 9.76RR45 pKa = 11.84PQWDD49 pKa = 3.9GRR51 pKa = 11.84PVPRR55 pKa = 11.84LARR58 pKa = 11.84AAVSYY63 pKa = 8.39LYY65 pKa = 10.6CDD67 pKa = 3.6VPVQVPLEE75 pKa = 4.3KK76 pKa = 10.51DD77 pKa = 3.04DD78 pKa = 3.78LTRR81 pKa = 11.84LLGYY85 pKa = 10.18LCEE88 pKa = 4.59HH89 pKa = 6.8LTMPPSLDD97 pKa = 3.23RR98 pKa = 11.84QTWVTDD104 pKa = 3.34KK105 pKa = 10.63KK106 pKa = 10.74RR107 pKa = 11.84AMEE110 pKa = 4.46AFPPKK115 pKa = 9.44RR116 pKa = 11.84RR117 pKa = 11.84AMALTKK123 pKa = 10.86ANIFLDD129 pKa = 4.14EE130 pKa = 4.18VARR133 pKa = 11.84DD134 pKa = 3.75LDD136 pKa = 3.24KK137 pKa = 11.55HH138 pKa = 5.23MPARR142 pKa = 11.84LLALLPYY149 pKa = 9.89LGKK152 pKa = 10.54LRR154 pKa = 11.84GVGATHH160 pKa = 7.06DD161 pKa = 3.52QATAFLLYY169 pKa = 10.65SSLLSDD175 pKa = 4.34HH176 pKa = 6.96CPLAAEE182 pKa = 4.28WGLFALTQPKK192 pKa = 8.85EE193 pKa = 4.08AKK195 pKa = 9.59EE196 pKa = 3.81VSNFLKK202 pKa = 10.81AVGANASSYY211 pKa = 10.86GALLVEE217 pKa = 4.54TDD219 pKa = 3.55TLLGRR224 pKa = 11.84DD225 pKa = 3.72TPGTDD230 pKa = 3.07LVADD234 pKa = 3.56SRR236 pKa = 11.84RR237 pKa = 11.84RR238 pKa = 11.84CDD240 pKa = 3.54LEE242 pKa = 3.89LARR245 pKa = 11.84EE246 pKa = 4.0EE247 pKa = 4.16VLAEE251 pKa = 4.17FDD253 pKa = 4.71DD254 pKa = 4.11SALRR258 pKa = 11.84SAVRR262 pKa = 11.84RR263 pKa = 11.84VLRR266 pKa = 11.84HH267 pKa = 5.33EE268 pKa = 4.29LRR270 pKa = 11.84SEE272 pKa = 3.82HH273 pKa = 5.41GSYY276 pKa = 10.31RR277 pKa = 11.84IEE279 pKa = 4.28YY280 pKa = 7.02PTQEE284 pKa = 4.28DD285 pKa = 3.6HH286 pKa = 5.84WTSRR290 pKa = 11.84WSWAVNGSHH299 pKa = 6.6SALLAGEE306 pKa = 4.25FPVPNEE312 pKa = 4.04VKK314 pKa = 10.15RR315 pKa = 11.84QVHH318 pKa = 5.55KK319 pKa = 9.87LHH321 pKa = 6.35RR322 pKa = 11.84RR323 pKa = 11.84AWLEE327 pKa = 3.89TIEE330 pKa = 4.46RR331 pKa = 11.84NPCIGWDD338 pKa = 3.02GKK340 pKa = 8.75TFVSCAPKK348 pKa = 10.27PEE350 pKa = 4.39CGKK353 pKa = 9.92IRR355 pKa = 11.84AIYY358 pKa = 10.33ACDD361 pKa = 3.68TINYY365 pKa = 8.97LAFEE369 pKa = 4.3HH370 pKa = 6.71LMAGVEE376 pKa = 4.24SRR378 pKa = 11.84WLGKK382 pKa = 10.09RR383 pKa = 11.84VILNPGRR390 pKa = 11.84GGHH393 pKa = 6.5LGIAEE398 pKa = 3.99RR399 pKa = 11.84VAACRR404 pKa = 11.84SRR406 pKa = 11.84SGISAMLDD414 pKa = 3.2YY415 pKa = 11.48DD416 pKa = 4.67DD417 pKa = 5.98FNSHH421 pKa = 5.95HH422 pKa = 5.91TTKK425 pKa = 10.93AMQILIEE432 pKa = 4.11EE433 pKa = 4.4TAALTGYY440 pKa = 9.61PPEE443 pKa = 4.15LTKK446 pKa = 10.66PLLDD450 pKa = 3.66SFEE453 pKa = 5.25KK454 pKa = 10.67EE455 pKa = 4.92DD456 pKa = 3.78IYY458 pKa = 11.45LDD460 pKa = 3.68GKK462 pKa = 10.81LVGRR466 pKa = 11.84VKK468 pKa = 9.49GTLMSGHH475 pKa = 6.99RR476 pKa = 11.84CTTYY480 pKa = 10.84VNSVLNMAYY489 pKa = 10.81LMVVLGEE496 pKa = 4.07DD497 pKa = 2.9WVMEE501 pKa = 4.17RR502 pKa = 11.84QSLHH506 pKa = 7.11VGDD509 pKa = 4.61DD510 pKa = 3.37VYY512 pKa = 11.02MGLKK516 pKa = 10.45NYY518 pKa = 10.77GEE520 pKa = 4.51TGWMISKK527 pKa = 10.28IMGSRR532 pKa = 11.84LRR534 pKa = 11.84MNRR537 pKa = 11.84RR538 pKa = 11.84KK539 pKa = 10.03QSVGHH544 pKa = 6.03VSTEE548 pKa = 3.89FLRR551 pKa = 11.84VASDD555 pKa = 3.18ARR557 pKa = 11.84DD558 pKa = 3.38SYY560 pKa = 11.92GYY562 pKa = 10.14LARR565 pKa = 11.84AAANLIAGNWYY576 pKa = 10.11SDD578 pKa = 3.18KK579 pKa = 10.97LLNPSEE585 pKa = 4.21GLITMIQGARR595 pKa = 11.84TLANRR600 pKa = 11.84ARR602 pKa = 11.84NGNAPLLLEE611 pKa = 4.3SAVKK615 pKa = 10.17RR616 pKa = 11.84VLGADD621 pKa = 3.89CPDD624 pKa = 4.23DD625 pKa = 3.39ATLRR629 pKa = 11.84HH630 pKa = 6.14MLTGSAAVNDD640 pKa = 4.53GPQYY644 pKa = 10.53LSSGRR649 pKa = 11.84HH650 pKa = 3.35RR651 pKa = 11.84RR652 pKa = 11.84VRR654 pKa = 11.84TSVVFEE660 pKa = 4.24VVDD663 pKa = 3.58TTGYY667 pKa = 8.73ATLPDD672 pKa = 4.34LASKK676 pKa = 10.78AYY678 pKa = 9.16LAKK681 pKa = 10.18HH682 pKa = 6.13ASPLEE687 pKa = 3.99IATLTSSGIDD697 pKa = 3.39PLPTMRR703 pKa = 11.84KK704 pKa = 9.3SSWTKK709 pKa = 10.73SLALSDD715 pKa = 4.33RR716 pKa = 11.84KK717 pKa = 10.61FEE719 pKa = 4.47TIVFKK724 pKa = 10.97PMEE727 pKa = 4.0QWPAVGSVQAEE738 pKa = 3.84HH739 pKa = 7.32LLRR742 pKa = 11.84EE743 pKa = 4.39RR744 pKa = 11.84KK745 pKa = 8.56PHH747 pKa = 6.0GVLLKK752 pKa = 10.9YY753 pKa = 10.14PLLVLMRR760 pKa = 11.84DD761 pKa = 4.47RR762 pKa = 11.84IPEE765 pKa = 3.91PVLRR769 pKa = 11.84VAVAAAGGRR778 pKa = 11.84DD779 pKa = 3.75YY780 pKa = 11.76VDD782 pKa = 5.79DD783 pKa = 4.54ISLEE787 pKa = 3.89AWGDD791 pKa = 3.57YY792 pKa = 11.05SNPTIVNTVLSYY804 pKa = 11.58SDD806 pKa = 4.55ASMLSKK812 pKa = 10.39RR813 pKa = 11.84ASVSVLTSRR822 pKa = 11.84RR823 pKa = 11.84RR824 pKa = 11.84CYY826 pKa = 10.4VV827 pKa = 2.75
Molecular weight: 92.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1599 |
772 |
827 |
799.5 |
86.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.633 ± 2.037 | 1.126 ± 0.233 |
5.691 ± 0.255 | 4.315 ± 0.808 |
2.877 ± 0.589 | 8.38 ± 1.241 |
2.064 ± 0.428 | 3.502 ± 0.17 |
3.002 ± 0.97 | 9.193 ± 1.907 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.064 ± 0.602 | 3.315 ± 0.383 |
6.942 ± 1.337 | 2.752 ± 0.5 |
7.067 ± 1.09 | 6.942 ± 0.225 |
6.504 ± 0.763 | 6.879 ± 0.096 |
1.438 ± 0.269 | 3.315 ± 0.138 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
