
Paracoccus sp. DMF
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Paracoccus; unclassified Paracoccus
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
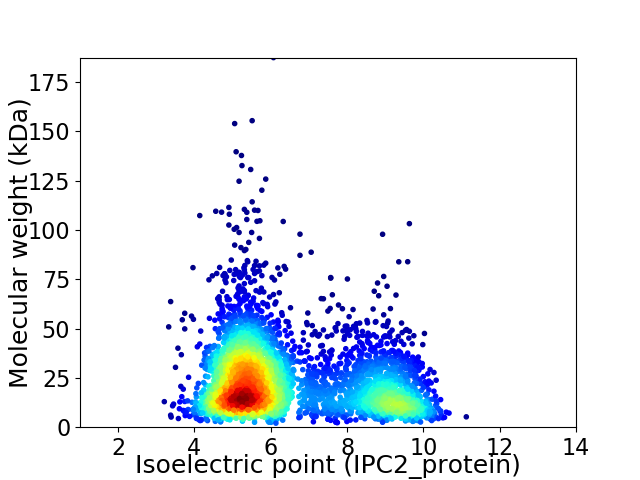
Virtual 2D-PAGE plot for 3948 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Y8NX12|A0A4Y8NX12_9RHOB Glycerol-3-phosphate acyltransferase OS=Paracoccus sp. DMF OX=400837 GN=plsY PE=3 SV=1
MM1 pKa = 7.44KK2 pKa = 10.19KK3 pKa = 10.41VLFATTALVMTAGVAAAEE21 pKa = 4.42VAVSGDD27 pKa = 2.72GRR29 pKa = 11.84MGMIYY34 pKa = 10.3DD35 pKa = 3.98GNDD38 pKa = 3.0AQFSSRR44 pKa = 11.84ARR46 pKa = 11.84VTFTLTGEE54 pKa = 4.16SDD56 pKa = 3.06AGLSFGGAFRR66 pKa = 11.84VDD68 pKa = 3.32QEE70 pKa = 4.01SDD72 pKa = 3.42YY73 pKa = 10.06TAGYY77 pKa = 9.85AGSEE81 pKa = 3.9RR82 pKa = 11.84SAARR86 pKa = 11.84GTAGAVWISGTYY98 pKa = 10.15GKK100 pKa = 10.88LSMGDD105 pKa = 3.25VVGAAEE111 pKa = 4.24AAIGDD116 pKa = 4.28LPEE119 pKa = 4.2VGYY122 pKa = 9.54TDD124 pKa = 4.57GEE126 pKa = 4.17FGGDD130 pKa = 3.07IEE132 pKa = 4.83EE133 pKa = 4.2INYY136 pKa = 10.09LVGDD140 pKa = 4.77GEE142 pKa = 5.12NEE144 pKa = 4.15DD145 pKa = 3.56QGPTILYY152 pKa = 8.67EE153 pKa = 3.97YY154 pKa = 9.14TVNNISFFASATDD167 pKa = 3.81GSNNSTYY174 pKa = 10.81GVAGSDD180 pKa = 3.43EE181 pKa = 4.85GEE183 pKa = 4.41DD184 pKa = 3.66VPSDD188 pKa = 3.44SVAYY192 pKa = 9.89SLAVKK197 pKa = 10.77YY198 pKa = 10.3EE199 pKa = 3.84GSNWWAALSYY209 pKa = 11.02SEE211 pKa = 5.31ADD213 pKa = 4.07VYY215 pKa = 11.68DD216 pKa = 4.97LDD218 pKa = 4.02PTSTSVTASASEE230 pKa = 3.83LALAGGASFNNFSVMATYY248 pKa = 10.82LKK250 pKa = 10.65YY251 pKa = 10.49DD252 pKa = 3.6DD253 pKa = 5.32RR254 pKa = 11.84FVSGPVLGDD263 pKa = 3.52DD264 pKa = 3.71EE265 pKa = 5.98FIVGGFAVDD274 pKa = 3.35ATLEE278 pKa = 4.12EE279 pKa = 4.68TIGLGATYY287 pKa = 10.68QMDD290 pKa = 5.34AILVEE295 pKa = 3.99GFYY298 pKa = 10.93RR299 pKa = 11.84KK300 pKa = 10.02DD301 pKa = 3.19KK302 pKa = 10.9YY303 pKa = 11.5DD304 pKa = 4.57LIGGGSEE311 pKa = 4.41SYY313 pKa = 11.3DD314 pKa = 3.44SFGIGADD321 pKa = 3.47YY322 pKa = 11.21DD323 pKa = 4.05LGGGAVLAGGIIDD336 pKa = 4.38TDD338 pKa = 3.96YY339 pKa = 11.85LDD341 pKa = 4.77DD342 pKa = 3.81TVADD346 pKa = 3.47IGVKK350 pKa = 10.55FKK352 pKa = 11.06FF353 pKa = 3.75
MM1 pKa = 7.44KK2 pKa = 10.19KK3 pKa = 10.41VLFATTALVMTAGVAAAEE21 pKa = 4.42VAVSGDD27 pKa = 2.72GRR29 pKa = 11.84MGMIYY34 pKa = 10.3DD35 pKa = 3.98GNDD38 pKa = 3.0AQFSSRR44 pKa = 11.84ARR46 pKa = 11.84VTFTLTGEE54 pKa = 4.16SDD56 pKa = 3.06AGLSFGGAFRR66 pKa = 11.84VDD68 pKa = 3.32QEE70 pKa = 4.01SDD72 pKa = 3.42YY73 pKa = 10.06TAGYY77 pKa = 9.85AGSEE81 pKa = 3.9RR82 pKa = 11.84SAARR86 pKa = 11.84GTAGAVWISGTYY98 pKa = 10.15GKK100 pKa = 10.88LSMGDD105 pKa = 3.25VVGAAEE111 pKa = 4.24AAIGDD116 pKa = 4.28LPEE119 pKa = 4.2VGYY122 pKa = 9.54TDD124 pKa = 4.57GEE126 pKa = 4.17FGGDD130 pKa = 3.07IEE132 pKa = 4.83EE133 pKa = 4.2INYY136 pKa = 10.09LVGDD140 pKa = 4.77GEE142 pKa = 5.12NEE144 pKa = 4.15DD145 pKa = 3.56QGPTILYY152 pKa = 8.67EE153 pKa = 3.97YY154 pKa = 9.14TVNNISFFASATDD167 pKa = 3.81GSNNSTYY174 pKa = 10.81GVAGSDD180 pKa = 3.43EE181 pKa = 4.85GEE183 pKa = 4.41DD184 pKa = 3.66VPSDD188 pKa = 3.44SVAYY192 pKa = 9.89SLAVKK197 pKa = 10.77YY198 pKa = 10.3EE199 pKa = 3.84GSNWWAALSYY209 pKa = 11.02SEE211 pKa = 5.31ADD213 pKa = 4.07VYY215 pKa = 11.68DD216 pKa = 4.97LDD218 pKa = 4.02PTSTSVTASASEE230 pKa = 3.83LALAGGASFNNFSVMATYY248 pKa = 10.82LKK250 pKa = 10.65YY251 pKa = 10.49DD252 pKa = 3.6DD253 pKa = 5.32RR254 pKa = 11.84FVSGPVLGDD263 pKa = 3.52DD264 pKa = 3.71EE265 pKa = 5.98FIVGGFAVDD274 pKa = 3.35ATLEE278 pKa = 4.12EE279 pKa = 4.68TIGLGATYY287 pKa = 10.68QMDD290 pKa = 5.34AILVEE295 pKa = 3.99GFYY298 pKa = 10.93RR299 pKa = 11.84KK300 pKa = 10.02DD301 pKa = 3.19KK302 pKa = 10.9YY303 pKa = 11.5DD304 pKa = 4.57LIGGGSEE311 pKa = 4.41SYY313 pKa = 11.3DD314 pKa = 3.44SFGIGADD321 pKa = 3.47YY322 pKa = 11.21DD323 pKa = 4.05LGGGAVLAGGIIDD336 pKa = 4.38TDD338 pKa = 3.96YY339 pKa = 11.85LDD341 pKa = 4.77DD342 pKa = 3.81TVADD346 pKa = 3.47IGVKK350 pKa = 10.55FKK352 pKa = 11.06FF353 pKa = 3.75
Molecular weight: 36.73 kDa
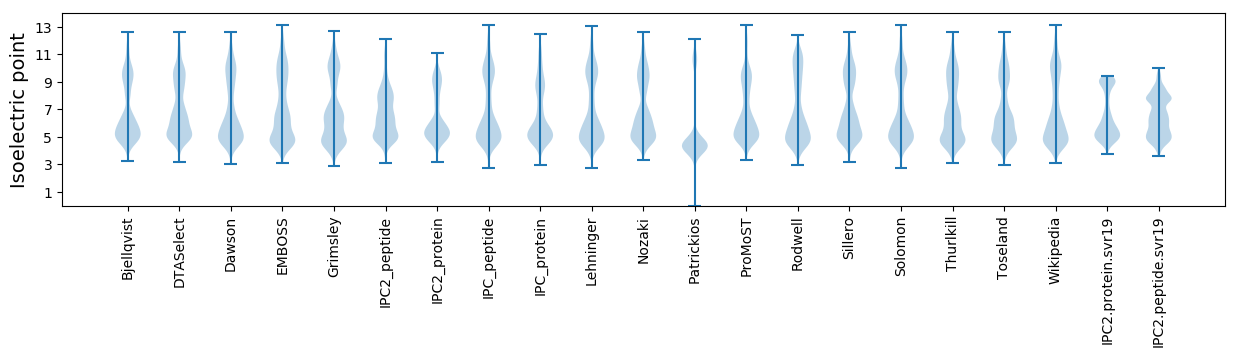
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Y8P5P5|A0A4Y8P5P5_9RHOB ABC-F family ATP-binding cassette domain-containing protein (Fragment) OS=Paracoccus sp. DMF OX=400837 GN=E3D03_01325 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.42RR3 pKa = 11.84TYY5 pKa = 10.34QPNKK9 pKa = 8.21RR10 pKa = 11.84KK11 pKa = 9.62RR12 pKa = 11.84SKK14 pKa = 9.59VHH16 pKa = 5.83GFRR19 pKa = 11.84SRR21 pKa = 11.84MSSPNGRR28 pKa = 11.84NVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.05GRR39 pKa = 11.84KK40 pKa = 8.7VLSAA44 pKa = 4.05
MM1 pKa = 7.35KK2 pKa = 9.42RR3 pKa = 11.84TYY5 pKa = 10.34QPNKK9 pKa = 8.21RR10 pKa = 11.84KK11 pKa = 9.62RR12 pKa = 11.84SKK14 pKa = 9.59VHH16 pKa = 5.83GFRR19 pKa = 11.84SRR21 pKa = 11.84MSSPNGRR28 pKa = 11.84NVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.05GRR39 pKa = 11.84KK40 pKa = 8.7VLSAA44 pKa = 4.05
Molecular weight: 5.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
905382 |
20 |
1702 |
229.3 |
25.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.336 ± 0.058 | 0.859 ± 0.012 |
5.679 ± 0.033 | 6.15 ± 0.043 |
3.892 ± 0.032 | 8.456 ± 0.037 |
2.06 ± 0.02 | 5.757 ± 0.034 |
3.93 ± 0.043 | 9.938 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.802 ± 0.02 | 2.949 ± 0.028 |
4.859 ± 0.03 | 3.356 ± 0.023 |
6.632 ± 0.049 | 5.246 ± 0.032 |
5.172 ± 0.036 | 6.971 ± 0.033 |
1.433 ± 0.021 | 2.52 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
