
Chimpanzee faeces associated microphage 3
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.71
Get precalculated fractions of proteins
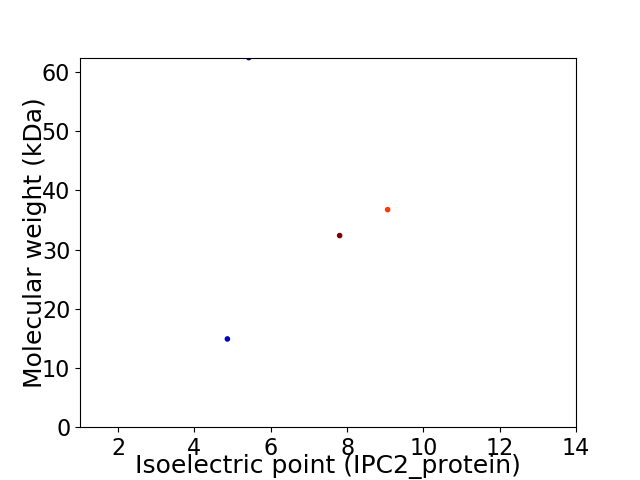
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A186YBY4|A0A186YBY4_9VIRU Minor CP 1 OS=Chimpanzee faeces associated microphage 3 OX=1676183 PE=4 SV=1
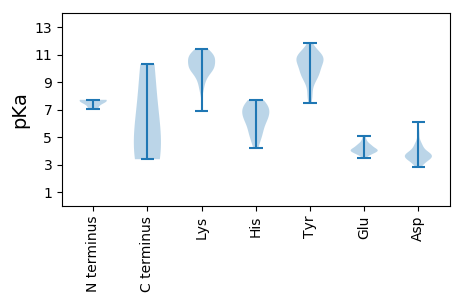
MM1 pKa = 7.63TFFTLFNKK9 pKa = 9.8PSPVGTACNTPSLTIQSEE27 pKa = 4.28KK28 pKa = 10.56AQCDD32 pKa = 3.02INAIIARR39 pKa = 11.84YY40 pKa = 9.21KK41 pKa = 8.51KK42 pKa = 9.09TGVVDD47 pKa = 4.5HH48 pKa = 7.23IKK50 pKa = 10.23RR51 pKa = 11.84DD52 pKa = 3.47QPLYY56 pKa = 10.8ADD58 pKa = 4.54CEE60 pKa = 4.07QAITDD65 pKa = 4.08LEE67 pKa = 4.16KK68 pKa = 11.12ARR70 pKa = 11.84ILVEE74 pKa = 4.04DD75 pKa = 3.98TEE77 pKa = 4.52EE78 pKa = 4.29AFWMLPSSVRR88 pKa = 11.84DD89 pKa = 3.78LIGEE93 pKa = 4.27PSNLPTWASVNRR105 pKa = 11.84SEE107 pKa = 4.26AEE109 pKa = 3.68KK110 pKa = 11.08YY111 pKa = 10.98GFIKK115 pKa = 10.02PQSTPPVDD123 pKa = 3.71SATPPPVDD131 pKa = 3.46SAANGG136 pKa = 3.39
MM1 pKa = 7.63TFFTLFNKK9 pKa = 9.8PSPVGTACNTPSLTIQSEE27 pKa = 4.28KK28 pKa = 10.56AQCDD32 pKa = 3.02INAIIARR39 pKa = 11.84YY40 pKa = 9.21KK41 pKa = 8.51KK42 pKa = 9.09TGVVDD47 pKa = 4.5HH48 pKa = 7.23IKK50 pKa = 10.23RR51 pKa = 11.84DD52 pKa = 3.47QPLYY56 pKa = 10.8ADD58 pKa = 4.54CEE60 pKa = 4.07QAITDD65 pKa = 4.08LEE67 pKa = 4.16KK68 pKa = 11.12ARR70 pKa = 11.84ILVEE74 pKa = 4.04DD75 pKa = 3.98TEE77 pKa = 4.52EE78 pKa = 4.29AFWMLPSSVRR88 pKa = 11.84DD89 pKa = 3.78LIGEE93 pKa = 4.27PSNLPTWASVNRR105 pKa = 11.84SEE107 pKa = 4.26AEE109 pKa = 3.68KK110 pKa = 11.08YY111 pKa = 10.98GFIKK115 pKa = 10.02PQSTPPVDD123 pKa = 3.71SATPPPVDD131 pKa = 3.46SAANGG136 pKa = 3.39
Molecular weight: 14.91 kDa
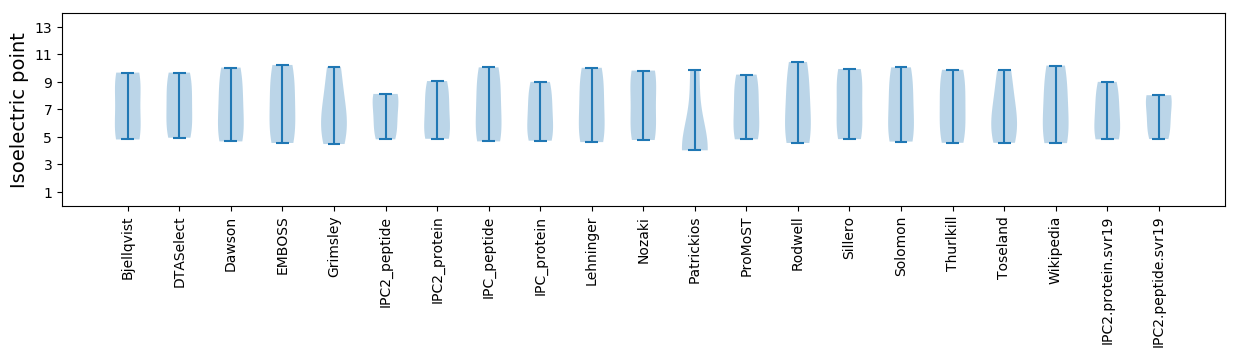
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A186YBY4|A0A186YBY4_9VIRU Minor CP 1 OS=Chimpanzee faeces associated microphage 3 OX=1676183 PE=4 SV=1
MM1 pKa = 7.62ARR3 pKa = 11.84PPPNWRR9 pKa = 11.84RR10 pKa = 11.84LFCLKK15 pKa = 10.16GDD17 pKa = 3.71YY18 pKa = 9.54MAFGVDD24 pKa = 3.48DD25 pKa = 4.43ALFAAVAGGSQGASNIAGAFLGDD48 pKa = 3.62YY49 pKa = 9.87FNRR52 pKa = 11.84KK53 pKa = 8.51AASKK57 pKa = 9.92QRR59 pKa = 11.84QASWDD64 pKa = 3.57MLLAEE69 pKa = 4.24QAFNARR75 pKa = 11.84EE76 pKa = 3.98AQKK79 pKa = 10.6NRR81 pKa = 11.84EE82 pKa = 3.94WQEE85 pKa = 3.82RR86 pKa = 11.84LSNTAHH92 pKa = 5.37QRR94 pKa = 11.84EE95 pKa = 4.52VADD98 pKa = 4.56LRR100 pKa = 11.84AAGLNPVLSATGGNGASTGSGAMATSNMSAPDD132 pKa = 4.02LSALAQSGQAFSRR145 pKa = 11.84VGNGVVDD152 pKa = 3.76KK153 pKa = 10.85ALRR156 pKa = 11.84AASLRR161 pKa = 11.84SDD163 pKa = 3.44IKK165 pKa = 11.28LMEE168 pKa = 4.86SNASNAEE175 pKa = 3.87AQAANSWEE183 pKa = 3.87NVLYY187 pKa = 10.44TKK189 pKa = 11.16ANTAKK194 pKa = 9.96VLAEE198 pKa = 4.0AGVFEE203 pKa = 4.38SQLDD207 pKa = 3.8RR208 pKa = 11.84IEE210 pKa = 3.94WLKK213 pKa = 11.14RR214 pKa = 11.84NSPNTWYY221 pKa = 10.64LWGDD225 pKa = 3.34KK226 pKa = 10.7DD227 pKa = 3.72PSVWQLLPGFSRR239 pKa = 11.84GSEE242 pKa = 3.94FGAQSVGNGVKK253 pKa = 9.88SVYY256 pKa = 10.77DD257 pKa = 3.61FVKK260 pKa = 10.73DD261 pKa = 4.53PIASAVKK268 pKa = 9.95KK269 pKa = 10.48VYY271 pKa = 10.6NAVKK275 pKa = 10.73DD276 pKa = 4.81FGANNASSSSNQSIGYY292 pKa = 9.29SSYY295 pKa = 11.85DD296 pKa = 3.31KK297 pKa = 11.15ASSSDD302 pKa = 3.58FGKK305 pKa = 10.86VPLTTTRR312 pKa = 11.84WRR314 pKa = 11.84KK315 pKa = 8.19GQPLRR320 pKa = 11.84SGKK323 pKa = 9.74FDD325 pKa = 3.5YY326 pKa = 11.27QEE328 pKa = 3.79VRR330 pKa = 11.84RR331 pKa = 11.84KK332 pKa = 10.04RR333 pKa = 11.84LEE335 pKa = 4.04SLKK338 pKa = 10.8KK339 pKa = 10.29
MM1 pKa = 7.62ARR3 pKa = 11.84PPPNWRR9 pKa = 11.84RR10 pKa = 11.84LFCLKK15 pKa = 10.16GDD17 pKa = 3.71YY18 pKa = 9.54MAFGVDD24 pKa = 3.48DD25 pKa = 4.43ALFAAVAGGSQGASNIAGAFLGDD48 pKa = 3.62YY49 pKa = 9.87FNRR52 pKa = 11.84KK53 pKa = 8.51AASKK57 pKa = 9.92QRR59 pKa = 11.84QASWDD64 pKa = 3.57MLLAEE69 pKa = 4.24QAFNARR75 pKa = 11.84EE76 pKa = 3.98AQKK79 pKa = 10.6NRR81 pKa = 11.84EE82 pKa = 3.94WQEE85 pKa = 3.82RR86 pKa = 11.84LSNTAHH92 pKa = 5.37QRR94 pKa = 11.84EE95 pKa = 4.52VADD98 pKa = 4.56LRR100 pKa = 11.84AAGLNPVLSATGGNGASTGSGAMATSNMSAPDD132 pKa = 4.02LSALAQSGQAFSRR145 pKa = 11.84VGNGVVDD152 pKa = 3.76KK153 pKa = 10.85ALRR156 pKa = 11.84AASLRR161 pKa = 11.84SDD163 pKa = 3.44IKK165 pKa = 11.28LMEE168 pKa = 4.86SNASNAEE175 pKa = 3.87AQAANSWEE183 pKa = 3.87NVLYY187 pKa = 10.44TKK189 pKa = 11.16ANTAKK194 pKa = 9.96VLAEE198 pKa = 4.0AGVFEE203 pKa = 4.38SQLDD207 pKa = 3.8RR208 pKa = 11.84IEE210 pKa = 3.94WLKK213 pKa = 11.14RR214 pKa = 11.84NSPNTWYY221 pKa = 10.64LWGDD225 pKa = 3.34KK226 pKa = 10.7DD227 pKa = 3.72PSVWQLLPGFSRR239 pKa = 11.84GSEE242 pKa = 3.94FGAQSVGNGVKK253 pKa = 9.88SVYY256 pKa = 10.77DD257 pKa = 3.61FVKK260 pKa = 10.73DD261 pKa = 4.53PIASAVKK268 pKa = 9.95KK269 pKa = 10.48VYY271 pKa = 10.6NAVKK275 pKa = 10.73DD276 pKa = 4.81FGANNASSSSNQSIGYY292 pKa = 9.29SSYY295 pKa = 11.85DD296 pKa = 3.31KK297 pKa = 11.15ASSSDD302 pKa = 3.58FGKK305 pKa = 10.86VPLTTTRR312 pKa = 11.84WRR314 pKa = 11.84KK315 pKa = 8.19GQPLRR320 pKa = 11.84SGKK323 pKa = 9.74FDD325 pKa = 3.5YY326 pKa = 11.27QEE328 pKa = 3.79VRR330 pKa = 11.84RR331 pKa = 11.84KK332 pKa = 10.04RR333 pKa = 11.84LEE335 pKa = 4.04SLKK338 pKa = 10.8KK339 pKa = 10.29
Molecular weight: 36.76 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1315 |
136 |
558 |
328.8 |
36.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.365 ± 2.031 | 1.597 ± 0.545 |
6.54 ± 0.42 | 4.411 ± 0.407 |
5.095 ± 0.427 | 7.072 ± 0.538 |
1.749 ± 0.594 | 3.27 ± 0.762 |
5.019 ± 0.606 | 8.213 ± 0.687 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.825 ± 0.078 | 5.171 ± 0.56 |
5.399 ± 0.778 | 3.422 ± 0.605 |
6.236 ± 0.726 | 9.658 ± 0.605 |
4.867 ± 0.896 | 6.312 ± 0.731 |
2.205 ± 0.151 | 3.574 ± 0.417 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
