
Rhodobacter blasticus DSM 2131
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Rhodobacter; Rhodobacter blasticus
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
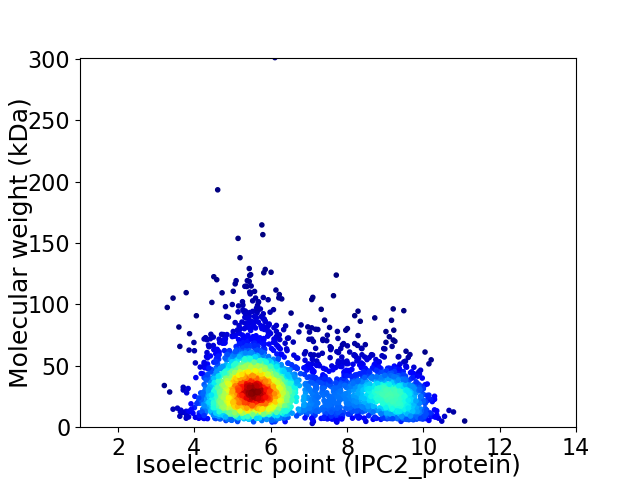
Virtual 2D-PAGE plot for 3451 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2T4J8Z6|A0A2T4J8Z6_RHOBL TonB-dependent receptor OS=Rhodobacter blasticus DSM 2131 OX=1188250 GN=C5F44_10070 PE=3 SV=1
MM1 pKa = 7.26MATFTVVPVVTNPATQKK18 pKa = 10.17LQGVINVTAGDD29 pKa = 4.04TYY31 pKa = 10.82IIDD34 pKa = 3.72PTVSVGITFNPPAGTPLGTTVSYY57 pKa = 10.92NIQINAANASAGTAQRR73 pKa = 11.84LITLGDD79 pKa = 3.34QTTANITIAPGVNAGRR95 pKa = 11.84YY96 pKa = 8.71DD97 pKa = 3.73FGGGTVDD104 pKa = 3.59GANYY108 pKa = 9.31TIGAGATTGDD118 pKa = 3.97LQLGDD123 pKa = 4.39GGNGSLTTVTSSVTLGDD140 pKa = 3.66GATVAGQITGGAGADD155 pKa = 3.56TLTVGTNVTVTEE167 pKa = 5.17AINLGAGAASGTFGTGFSAQSFAASGAGAKK197 pKa = 9.37TFDD200 pKa = 4.2FSNTTTFTGTGASVSIASTNAPVSVDD226 pKa = 3.82FDD228 pKa = 5.52DD229 pKa = 4.75NLSTNGTISVSSSTGAATLDD249 pKa = 2.79IGANLKK255 pKa = 8.92TGNATLHH262 pKa = 5.77TGGAISVVGGGTGSANMLTIGLDD285 pKa = 3.4AQVNGAITINGSGGTNTLTIGDD307 pKa = 3.93SSEE310 pKa = 4.42VYY312 pKa = 10.73GFIYY316 pKa = 10.75ANGTNSAATVTIGDD330 pKa = 4.6LSTVTGDD337 pKa = 3.2IYY339 pKa = 11.47ANSGGTLALGNIDD352 pKa = 3.42NSVTVGDD359 pKa = 4.05GTTTGSIFVNGLYY372 pKa = 10.25SDD374 pKa = 3.76NTVSVGLASTVGNLYY389 pKa = 10.43FGTDD393 pKa = 3.5GTVSQTDD400 pKa = 3.62PTSGQVLTMADD411 pKa = 3.36NSTAGLIYY419 pKa = 9.91LTASNSTFDD428 pKa = 3.48VTFGTGVTTNAIYY441 pKa = 9.98GYY443 pKa = 10.48GYY445 pKa = 10.62NSDD448 pKa = 4.27YY449 pKa = 11.07NVSIPDD455 pKa = 3.83DD456 pKa = 3.85FTANGYY462 pKa = 10.58VYY464 pKa = 10.57FGSTNSTTDD473 pKa = 3.01VTIGNNAVINTGALDD488 pKa = 3.44ISGSDD493 pKa = 3.34VTTLIGDD500 pKa = 4.26GLSVQYY506 pKa = 9.85FASFGDD512 pKa = 3.91LNGATDD518 pKa = 3.5VTIGQGANIGGALYY532 pKa = 10.95ANGSTVDD539 pKa = 4.15FTAGPDD545 pKa = 3.68FDD547 pKa = 4.39VSGYY551 pKa = 11.44AYY553 pKa = 10.91LNDD556 pKa = 3.71ANATYY561 pKa = 9.65TIDD564 pKa = 3.74IGPGSDD570 pKa = 2.71IGGGFYY576 pKa = 11.1ANGFNSDD583 pKa = 3.45VTLTTGDD590 pKa = 3.59NFIVSLDD597 pKa = 3.65AYY599 pKa = 10.01IGGTDD604 pKa = 3.39GTTTVTLGSGSSIGTVLNVGGDD626 pKa = 3.89TVNFTAGTGLTTGGAVTFGDD646 pKa = 3.95AVSLSTTVNLTGPTVFGGLLDD667 pKa = 3.85VNGDD671 pKa = 3.61QIIFNGGPNLTVNGPAYY688 pKa = 7.9FTNDD692 pKa = 3.19GGTSTIVLGDD702 pKa = 3.31NTTINGLADD711 pKa = 4.32FAAPNQDD718 pKa = 2.66VSLTIGTNVTIGTIYY733 pKa = 11.01GDD735 pKa = 3.59TTGTGSFNMTTGDD748 pKa = 4.27DD749 pKa = 3.5FTFANFYY756 pKa = 11.13GGTTDD761 pKa = 3.53NDD763 pKa = 3.33EE764 pKa = 4.38HH765 pKa = 5.71VTIGKK770 pKa = 8.27NWTMTGSGVYY780 pKa = 10.0DD781 pKa = 3.79AGTGNDD787 pKa = 3.65TLILGTTADD796 pKa = 3.99RR797 pKa = 11.84SSTNYY802 pKa = 9.91IYY804 pKa = 10.9GGEE807 pKa = 4.2SANDD811 pKa = 3.27NDD813 pKa = 4.01VLTFRR818 pKa = 11.84LNAADD823 pKa = 3.49QASFTTAALAAGWTLNPDD841 pKa = 3.53GTFKK845 pKa = 8.57TTNGAGAHH853 pKa = 6.42IQLTWQGQVINYY865 pKa = 7.03WEE867 pKa = 4.21SVAFCFAQGTEE878 pKa = 3.75ISTDD882 pKa = 3.11AGPVKK887 pKa = 10.88VEE889 pKa = 4.02DD890 pKa = 4.17LRR892 pKa = 11.84PGSMVLTMDD901 pKa = 4.49RR902 pKa = 11.84GYY904 pKa = 11.0QPVAWIGGRR913 pKa = 11.84QLSGADD919 pKa = 3.64LQRR922 pKa = 11.84KK923 pKa = 8.18PNLRR927 pKa = 11.84PIRR930 pKa = 11.84IAAGALGHH938 pKa = 6.44GLPTADD944 pKa = 4.13LVVSPQHH951 pKa = 5.72RR952 pKa = 11.84VLVRR956 pKa = 11.84SRR958 pKa = 11.84LSEE961 pKa = 3.83RR962 pKa = 11.84MFSDD966 pKa = 3.69RR967 pKa = 11.84EE968 pKa = 3.78VLIAAKK974 pKa = 10.11HH975 pKa = 4.81LVGLPGITVAEE986 pKa = 4.22EE987 pKa = 3.97MDD989 pKa = 4.02GVSYY993 pKa = 9.36WHH995 pKa = 6.62FLFEE999 pKa = 3.89QHH1001 pKa = 6.78EE1002 pKa = 4.54IVYY1005 pKa = 10.9SNGAPTEE1012 pKa = 4.2SLFTGPEE1019 pKa = 3.72ALKK1022 pKa = 10.88ALTPDD1027 pKa = 3.31QQQEE1031 pKa = 4.07ILTLFPEE1038 pKa = 4.81LSEE1041 pKa = 4.05TDD1043 pKa = 3.17AVLNGDD1049 pKa = 3.84RR1050 pKa = 11.84LIPARR1055 pKa = 11.84TIVPGRR1061 pKa = 11.84LARR1064 pKa = 11.84SFTRR1068 pKa = 11.84RR1069 pKa = 11.84ATKK1072 pKa = 10.32RR1073 pKa = 11.84GASDD1077 pKa = 3.2VLSPVV1082 pKa = 3.02
MM1 pKa = 7.26MATFTVVPVVTNPATQKK18 pKa = 10.17LQGVINVTAGDD29 pKa = 4.04TYY31 pKa = 10.82IIDD34 pKa = 3.72PTVSVGITFNPPAGTPLGTTVSYY57 pKa = 10.92NIQINAANASAGTAQRR73 pKa = 11.84LITLGDD79 pKa = 3.34QTTANITIAPGVNAGRR95 pKa = 11.84YY96 pKa = 8.71DD97 pKa = 3.73FGGGTVDD104 pKa = 3.59GANYY108 pKa = 9.31TIGAGATTGDD118 pKa = 3.97LQLGDD123 pKa = 4.39GGNGSLTTVTSSVTLGDD140 pKa = 3.66GATVAGQITGGAGADD155 pKa = 3.56TLTVGTNVTVTEE167 pKa = 5.17AINLGAGAASGTFGTGFSAQSFAASGAGAKK197 pKa = 9.37TFDD200 pKa = 4.2FSNTTTFTGTGASVSIASTNAPVSVDD226 pKa = 3.82FDD228 pKa = 5.52DD229 pKa = 4.75NLSTNGTISVSSSTGAATLDD249 pKa = 2.79IGANLKK255 pKa = 8.92TGNATLHH262 pKa = 5.77TGGAISVVGGGTGSANMLTIGLDD285 pKa = 3.4AQVNGAITINGSGGTNTLTIGDD307 pKa = 3.93SSEE310 pKa = 4.42VYY312 pKa = 10.73GFIYY316 pKa = 10.75ANGTNSAATVTIGDD330 pKa = 4.6LSTVTGDD337 pKa = 3.2IYY339 pKa = 11.47ANSGGTLALGNIDD352 pKa = 3.42NSVTVGDD359 pKa = 4.05GTTTGSIFVNGLYY372 pKa = 10.25SDD374 pKa = 3.76NTVSVGLASTVGNLYY389 pKa = 10.43FGTDD393 pKa = 3.5GTVSQTDD400 pKa = 3.62PTSGQVLTMADD411 pKa = 3.36NSTAGLIYY419 pKa = 9.91LTASNSTFDD428 pKa = 3.48VTFGTGVTTNAIYY441 pKa = 9.98GYY443 pKa = 10.48GYY445 pKa = 10.62NSDD448 pKa = 4.27YY449 pKa = 11.07NVSIPDD455 pKa = 3.83DD456 pKa = 3.85FTANGYY462 pKa = 10.58VYY464 pKa = 10.57FGSTNSTTDD473 pKa = 3.01VTIGNNAVINTGALDD488 pKa = 3.44ISGSDD493 pKa = 3.34VTTLIGDD500 pKa = 4.26GLSVQYY506 pKa = 9.85FASFGDD512 pKa = 3.91LNGATDD518 pKa = 3.5VTIGQGANIGGALYY532 pKa = 10.95ANGSTVDD539 pKa = 4.15FTAGPDD545 pKa = 3.68FDD547 pKa = 4.39VSGYY551 pKa = 11.44AYY553 pKa = 10.91LNDD556 pKa = 3.71ANATYY561 pKa = 9.65TIDD564 pKa = 3.74IGPGSDD570 pKa = 2.71IGGGFYY576 pKa = 11.1ANGFNSDD583 pKa = 3.45VTLTTGDD590 pKa = 3.59NFIVSLDD597 pKa = 3.65AYY599 pKa = 10.01IGGTDD604 pKa = 3.39GTTTVTLGSGSSIGTVLNVGGDD626 pKa = 3.89TVNFTAGTGLTTGGAVTFGDD646 pKa = 3.95AVSLSTTVNLTGPTVFGGLLDD667 pKa = 3.85VNGDD671 pKa = 3.61QIIFNGGPNLTVNGPAYY688 pKa = 7.9FTNDD692 pKa = 3.19GGTSTIVLGDD702 pKa = 3.31NTTINGLADD711 pKa = 4.32FAAPNQDD718 pKa = 2.66VSLTIGTNVTIGTIYY733 pKa = 11.01GDD735 pKa = 3.59TTGTGSFNMTTGDD748 pKa = 4.27DD749 pKa = 3.5FTFANFYY756 pKa = 11.13GGTTDD761 pKa = 3.53NDD763 pKa = 3.33EE764 pKa = 4.38HH765 pKa = 5.71VTIGKK770 pKa = 8.27NWTMTGSGVYY780 pKa = 10.0DD781 pKa = 3.79AGTGNDD787 pKa = 3.65TLILGTTADD796 pKa = 3.99RR797 pKa = 11.84SSTNYY802 pKa = 9.91IYY804 pKa = 10.9GGEE807 pKa = 4.2SANDD811 pKa = 3.27NDD813 pKa = 4.01VLTFRR818 pKa = 11.84LNAADD823 pKa = 3.49QASFTTAALAAGWTLNPDD841 pKa = 3.53GTFKK845 pKa = 8.57TTNGAGAHH853 pKa = 6.42IQLTWQGQVINYY865 pKa = 7.03WEE867 pKa = 4.21SVAFCFAQGTEE878 pKa = 3.75ISTDD882 pKa = 3.11AGPVKK887 pKa = 10.88VEE889 pKa = 4.02DD890 pKa = 4.17LRR892 pKa = 11.84PGSMVLTMDD901 pKa = 4.49RR902 pKa = 11.84GYY904 pKa = 11.0QPVAWIGGRR913 pKa = 11.84QLSGADD919 pKa = 3.64LQRR922 pKa = 11.84KK923 pKa = 8.18PNLRR927 pKa = 11.84PIRR930 pKa = 11.84IAAGALGHH938 pKa = 6.44GLPTADD944 pKa = 4.13LVVSPQHH951 pKa = 5.72RR952 pKa = 11.84VLVRR956 pKa = 11.84SRR958 pKa = 11.84LSEE961 pKa = 3.83RR962 pKa = 11.84MFSDD966 pKa = 3.69RR967 pKa = 11.84EE968 pKa = 3.78VLIAAKK974 pKa = 10.11HH975 pKa = 4.81LVGLPGITVAEE986 pKa = 4.22EE987 pKa = 3.97MDD989 pKa = 4.02GVSYY993 pKa = 9.36WHH995 pKa = 6.62FLFEE999 pKa = 3.89QHH1001 pKa = 6.78EE1002 pKa = 4.54IVYY1005 pKa = 10.9SNGAPTEE1012 pKa = 4.2SLFTGPEE1019 pKa = 3.72ALKK1022 pKa = 10.88ALTPDD1027 pKa = 3.31QQQEE1031 pKa = 4.07ILTLFPEE1038 pKa = 4.81LSEE1041 pKa = 4.05TDD1043 pKa = 3.17AVLNGDD1049 pKa = 3.84RR1050 pKa = 11.84LIPARR1055 pKa = 11.84TIVPGRR1061 pKa = 11.84LARR1064 pKa = 11.84SFTRR1068 pKa = 11.84RR1069 pKa = 11.84ATKK1072 pKa = 10.32RR1073 pKa = 11.84GASDD1077 pKa = 3.2VLSPVV1082 pKa = 3.02
Molecular weight: 109.57 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2T4J6X9|A0A2T4J6X9_RHOBL Transcriptional regulator OS=Rhodobacter blasticus DSM 2131 OX=1188250 GN=C5F44_13570 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 10.31QPSTLVRR12 pKa = 11.84KK13 pKa = 9.36RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.48GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.34GGRR28 pKa = 11.84LVLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.71GRR39 pKa = 11.84KK40 pKa = 8.73VLSAA44 pKa = 4.05
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 10.31QPSTLVRR12 pKa = 11.84KK13 pKa = 9.36RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.48GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.34GGRR28 pKa = 11.84LVLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.71GRR39 pKa = 11.84KK40 pKa = 8.73VLSAA44 pKa = 4.05
Molecular weight: 5.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1074197 |
28 |
2804 |
311.3 |
33.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.784 ± 0.068 | 0.853 ± 0.012 |
5.692 ± 0.034 | 5.363 ± 0.038 |
3.538 ± 0.026 | 9.09 ± 0.043 |
1.981 ± 0.021 | 4.614 ± 0.028 |
2.843 ± 0.035 | 10.643 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.617 ± 0.022 | 2.205 ± 0.025 |
5.674 ± 0.036 | 2.99 ± 0.019 |
7.206 ± 0.041 | 4.585 ± 0.027 |
5.376 ± 0.026 | 7.451 ± 0.037 |
1.473 ± 0.017 | 2.022 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
