
Capybara microvirus Cap1_SP_147
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.41
Get precalculated fractions of proteins
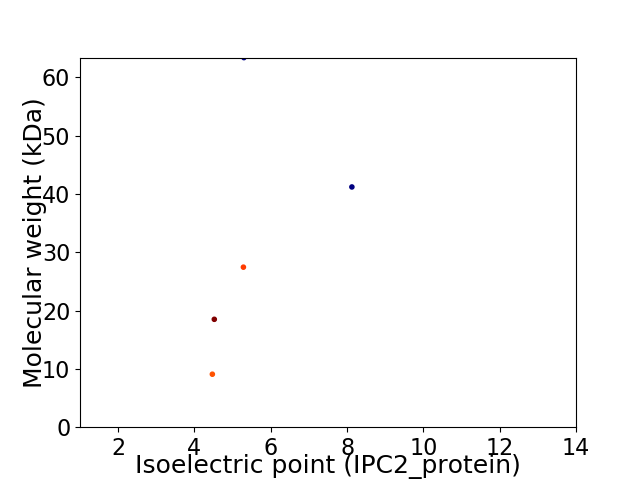
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
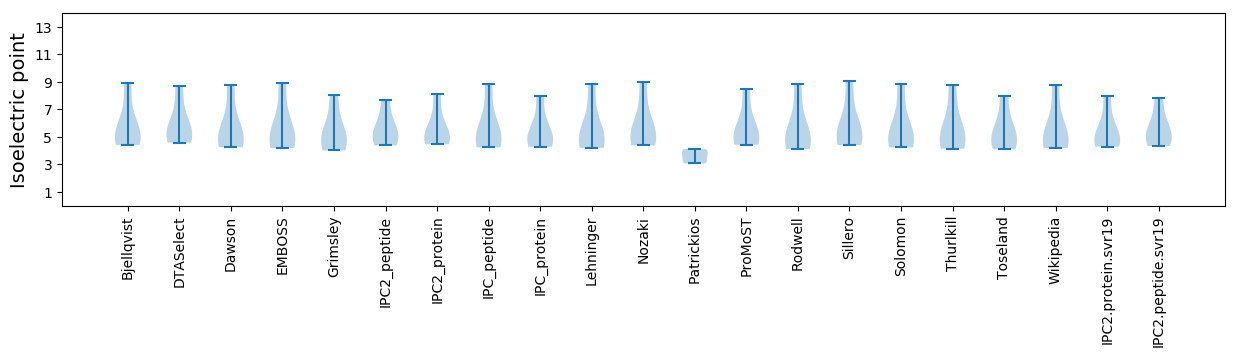
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W4E6|A0A4P8W4E6_9VIRU Minor capsid protein OS=Capybara microvirus Cap1_SP_147 OX=2585391 PE=4 SV=1
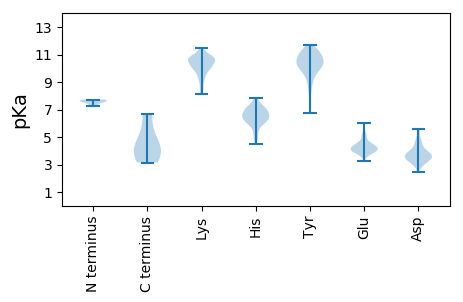
MM1 pKa = 7.59KK2 pKa = 10.63YY3 pKa = 9.96EE4 pKa = 4.08VYY6 pKa = 10.39AVKK9 pKa = 10.71DD10 pKa = 3.76SITGFNQPACEE21 pKa = 4.06LNQAAALRR29 pKa = 11.84SFKK32 pKa = 11.02NLINLNEE39 pKa = 4.07AFRR42 pKa = 11.84INAGDD47 pKa = 3.53YY48 pKa = 10.97SLFKK52 pKa = 10.65IADD55 pKa = 3.56FDD57 pKa = 4.3TDD59 pKa = 2.93TGYY62 pKa = 8.66CTGIDD67 pKa = 3.35PVFIVSGSSVLEE79 pKa = 4.1VNNAA83 pKa = 3.15
MM1 pKa = 7.59KK2 pKa = 10.63YY3 pKa = 9.96EE4 pKa = 4.08VYY6 pKa = 10.39AVKK9 pKa = 10.71DD10 pKa = 3.76SITGFNQPACEE21 pKa = 4.06LNQAAALRR29 pKa = 11.84SFKK32 pKa = 11.02NLINLNEE39 pKa = 4.07AFRR42 pKa = 11.84INAGDD47 pKa = 3.53YY48 pKa = 10.97SLFKK52 pKa = 10.65IADD55 pKa = 3.56FDD57 pKa = 4.3TDD59 pKa = 2.93TGYY62 pKa = 8.66CTGIDD67 pKa = 3.35PVFIVSGSSVLEE79 pKa = 4.1VNNAA83 pKa = 3.15
Molecular weight: 9.09 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W7Z9|A0A4P8W7Z9_9VIRU Major capsid protein OS=Capybara microvirus Cap1_SP_147 OX=2585391 PE=3 SV=1
MM1 pKa = 7.73CKK3 pKa = 10.08HH4 pKa = 6.35PLKK7 pKa = 10.53AFQYY11 pKa = 9.95GLHH14 pKa = 6.33EE15 pKa = 4.78KK16 pKa = 10.16SGKK19 pKa = 8.64PNYY22 pKa = 9.77IIAPYY27 pKa = 7.32QCKK30 pKa = 10.66YY31 pKa = 10.29IDD33 pKa = 3.79VDD35 pKa = 3.36IHH37 pKa = 6.73DD38 pKa = 5.2VPHH41 pKa = 6.64KK42 pKa = 10.09MYY44 pKa = 10.54NEE46 pKa = 4.05PVRR49 pKa = 11.84SQLTKK54 pKa = 10.82SIITSFLEE62 pKa = 4.32IPCGKK67 pKa = 10.18CIEE70 pKa = 4.44CRR72 pKa = 11.84LTYY75 pKa = 10.36ARR77 pKa = 11.84QWANRR82 pKa = 11.84CQVEE86 pKa = 3.66ADD88 pKa = 3.53QYY90 pKa = 11.37EE91 pKa = 4.56KK92 pKa = 11.36NCFITLTYY100 pKa = 10.4SDD102 pKa = 5.44DD103 pKa = 4.14YY104 pKa = 11.68IHH106 pKa = 7.04FNTFTDD112 pKa = 4.0PVTGEE117 pKa = 4.19LKK119 pKa = 10.61KK120 pKa = 11.11SPTLVKK126 pKa = 10.24RR127 pKa = 11.84DD128 pKa = 3.16IQNFMKK134 pKa = 10.71RR135 pKa = 11.84LRR137 pKa = 11.84KK138 pKa = 9.81KK139 pKa = 8.52YY140 pKa = 9.3GAGIRR145 pKa = 11.84FFACGEE151 pKa = 4.14YY152 pKa = 11.08GEE154 pKa = 4.73TTHH157 pKa = 6.99RR158 pKa = 11.84PHH160 pKa = 6.02YY161 pKa = 10.39HH162 pKa = 7.1LILFNHH168 pKa = 7.03DD169 pKa = 3.71FNDD172 pKa = 3.33KK173 pKa = 10.78KK174 pKa = 10.81LFKK177 pKa = 10.45VSRR180 pKa = 11.84GNPYY184 pKa = 10.26YY185 pKa = 10.18ISDD188 pKa = 4.26EE189 pKa = 5.06LSDD192 pKa = 3.64LWPFGFSMIGDD203 pKa = 3.97VSWQSCNYY211 pKa = 7.18VARR214 pKa = 11.84YY215 pKa = 6.75ITKK218 pKa = 9.87KK219 pKa = 10.12QYY221 pKa = 10.32KK222 pKa = 9.23KK223 pKa = 10.73KK224 pKa = 10.56NEE226 pKa = 3.96EE227 pKa = 3.95KK228 pKa = 10.78DD229 pKa = 3.84CIIYY233 pKa = 10.37NYY235 pKa = 9.9EE236 pKa = 3.9PEE238 pKa = 4.15FVLMSRR244 pKa = 11.84KK245 pKa = 9.53PGIGLKK251 pKa = 10.43YY252 pKa = 10.97YY253 pKa = 10.36EE254 pKa = 5.27DD255 pKa = 3.62NKK257 pKa = 10.73QDD259 pKa = 4.59FIRR262 pKa = 11.84FNGVYY267 pKa = 9.53LQNGRR272 pKa = 11.84LFTPPRR278 pKa = 11.84YY279 pKa = 9.29FNKK282 pKa = 10.3KK283 pKa = 10.18LEE285 pKa = 3.98IDD287 pKa = 3.86FGSEE291 pKa = 3.74YY292 pKa = 10.88LEE294 pKa = 3.98KK295 pKa = 10.64KK296 pKa = 9.53EE297 pKa = 3.9RR298 pKa = 11.84NKK300 pKa = 10.89KK301 pKa = 9.42IFNDD305 pKa = 3.38QSILKK310 pKa = 10.61DD311 pKa = 3.44EE312 pKa = 4.43LTDD315 pKa = 3.96LNYY318 pKa = 10.74LEE320 pKa = 4.57RR321 pKa = 11.84LQVEE325 pKa = 4.11EE326 pKa = 6.02DD327 pKa = 3.17ILLKK331 pKa = 9.27KK332 pKa = 9.24TKK334 pKa = 9.69ILHH337 pKa = 6.51RR338 pKa = 11.84NGADD342 pKa = 3.38PLGVLL347 pKa = 4.63
MM1 pKa = 7.73CKK3 pKa = 10.08HH4 pKa = 6.35PLKK7 pKa = 10.53AFQYY11 pKa = 9.95GLHH14 pKa = 6.33EE15 pKa = 4.78KK16 pKa = 10.16SGKK19 pKa = 8.64PNYY22 pKa = 9.77IIAPYY27 pKa = 7.32QCKK30 pKa = 10.66YY31 pKa = 10.29IDD33 pKa = 3.79VDD35 pKa = 3.36IHH37 pKa = 6.73DD38 pKa = 5.2VPHH41 pKa = 6.64KK42 pKa = 10.09MYY44 pKa = 10.54NEE46 pKa = 4.05PVRR49 pKa = 11.84SQLTKK54 pKa = 10.82SIITSFLEE62 pKa = 4.32IPCGKK67 pKa = 10.18CIEE70 pKa = 4.44CRR72 pKa = 11.84LTYY75 pKa = 10.36ARR77 pKa = 11.84QWANRR82 pKa = 11.84CQVEE86 pKa = 3.66ADD88 pKa = 3.53QYY90 pKa = 11.37EE91 pKa = 4.56KK92 pKa = 11.36NCFITLTYY100 pKa = 10.4SDD102 pKa = 5.44DD103 pKa = 4.14YY104 pKa = 11.68IHH106 pKa = 7.04FNTFTDD112 pKa = 4.0PVTGEE117 pKa = 4.19LKK119 pKa = 10.61KK120 pKa = 11.11SPTLVKK126 pKa = 10.24RR127 pKa = 11.84DD128 pKa = 3.16IQNFMKK134 pKa = 10.71RR135 pKa = 11.84LRR137 pKa = 11.84KK138 pKa = 9.81KK139 pKa = 8.52YY140 pKa = 9.3GAGIRR145 pKa = 11.84FFACGEE151 pKa = 4.14YY152 pKa = 11.08GEE154 pKa = 4.73TTHH157 pKa = 6.99RR158 pKa = 11.84PHH160 pKa = 6.02YY161 pKa = 10.39HH162 pKa = 7.1LILFNHH168 pKa = 7.03DD169 pKa = 3.71FNDD172 pKa = 3.33KK173 pKa = 10.78KK174 pKa = 10.81LFKK177 pKa = 10.45VSRR180 pKa = 11.84GNPYY184 pKa = 10.26YY185 pKa = 10.18ISDD188 pKa = 4.26EE189 pKa = 5.06LSDD192 pKa = 3.64LWPFGFSMIGDD203 pKa = 3.97VSWQSCNYY211 pKa = 7.18VARR214 pKa = 11.84YY215 pKa = 6.75ITKK218 pKa = 9.87KK219 pKa = 10.12QYY221 pKa = 10.32KK222 pKa = 9.23KK223 pKa = 10.73KK224 pKa = 10.56NEE226 pKa = 3.96EE227 pKa = 3.95KK228 pKa = 10.78DD229 pKa = 3.84CIIYY233 pKa = 10.37NYY235 pKa = 9.9EE236 pKa = 3.9PEE238 pKa = 4.15FVLMSRR244 pKa = 11.84KK245 pKa = 9.53PGIGLKK251 pKa = 10.43YY252 pKa = 10.97YY253 pKa = 10.36EE254 pKa = 5.27DD255 pKa = 3.62NKK257 pKa = 10.73QDD259 pKa = 4.59FIRR262 pKa = 11.84FNGVYY267 pKa = 9.53LQNGRR272 pKa = 11.84LFTPPRR278 pKa = 11.84YY279 pKa = 9.29FNKK282 pKa = 10.3KK283 pKa = 10.18LEE285 pKa = 3.98IDD287 pKa = 3.86FGSEE291 pKa = 3.74YY292 pKa = 10.88LEE294 pKa = 3.98KK295 pKa = 10.64KK296 pKa = 9.53EE297 pKa = 3.9RR298 pKa = 11.84NKK300 pKa = 10.89KK301 pKa = 9.42IFNDD305 pKa = 3.38QSILKK310 pKa = 10.61DD311 pKa = 3.44EE312 pKa = 4.43LTDD315 pKa = 3.96LNYY318 pKa = 10.74LEE320 pKa = 4.57RR321 pKa = 11.84LQVEE325 pKa = 4.11EE326 pKa = 6.02DD327 pKa = 3.17ILLKK331 pKa = 9.27KK332 pKa = 9.24TKK334 pKa = 9.69ILHH337 pKa = 6.51RR338 pKa = 11.84NGADD342 pKa = 3.38PLGVLL347 pKa = 4.63
Molecular weight: 41.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1419 |
83 |
567 |
283.8 |
31.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.906 ± 2.701 | 1.268 ± 0.57 |
5.708 ± 0.82 | 5.426 ± 0.625 |
5.004 ± 0.686 | 6.413 ± 0.672 |
2.044 ± 0.346 | 5.567 ± 0.545 |
5.99 ± 1.47 | 7.752 ± 0.361 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.396 ± 0.358 | 7.541 ± 0.94 |
5.004 ± 1.156 | 3.946 ± 0.338 |
3.453 ± 0.724 | 7.893 ± 1.672 |
6.131 ± 0.691 | 4.933 ± 0.86 |
1.198 ± 0.446 | 5.426 ± 0.733 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
