
Ctenophore-associated circular virus 3
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 8.02
Get precalculated fractions of proteins
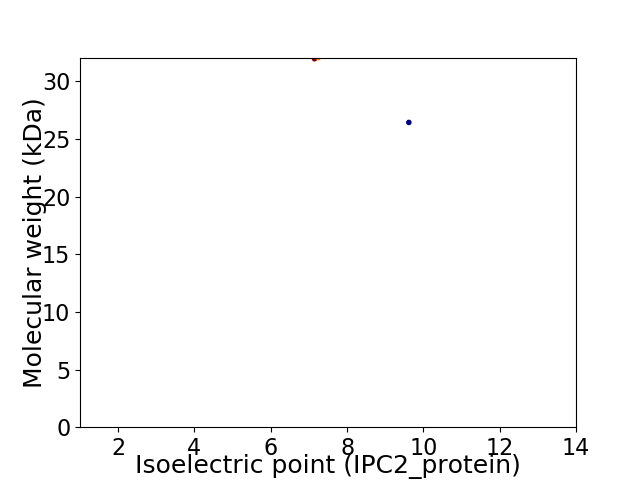
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A141MJB0|A0A141MJB0_9VIRU Uncharacterized protein OS=Ctenophore-associated circular virus 3 OX=1778560 PE=4 SV=1
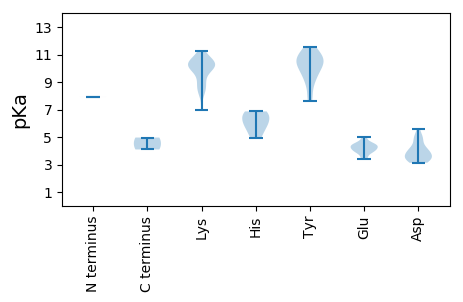
MM1 pKa = 7.91PGYY4 pKa = 9.29QFRR7 pKa = 11.84SATFTQFNVEE17 pKa = 4.18KK18 pKa = 10.62APEE21 pKa = 4.47LGNLQYY27 pKa = 10.78IAFGLEE33 pKa = 4.31TCPSTQRR40 pKa = 11.84QHH42 pKa = 4.93WQGFAYY48 pKa = 10.21SYY50 pKa = 11.3TKK52 pKa = 8.21MTHH55 pKa = 5.52KK56 pKa = 10.1KK57 pKa = 8.59WRR59 pKa = 11.84EE60 pKa = 3.95TFPGAHH66 pKa = 5.97IEE68 pKa = 4.06EE69 pKa = 4.45MKK71 pKa = 11.19GNFRR75 pKa = 11.84QNEE78 pKa = 4.69VYY80 pKa = 10.2CSKK83 pKa = 10.7EE84 pKa = 3.51GTYY87 pKa = 10.1TEE89 pKa = 5.02FGIKK93 pKa = 10.02PMEE96 pKa = 4.42NGKK99 pKa = 9.73KK100 pKa = 9.94RR101 pKa = 11.84SLAEE105 pKa = 3.87MKK107 pKa = 10.23EE108 pKa = 3.86QLDD111 pKa = 3.71NGEE114 pKa = 4.48KK115 pKa = 9.72PVEE118 pKa = 4.13VATKK122 pKa = 9.07SDD124 pKa = 3.15KK125 pKa = 10.88MFGTFLQYY133 pKa = 10.62RR134 pKa = 11.84SGLNEE139 pKa = 3.42YY140 pKa = 9.88AQYY143 pKa = 10.58IRR145 pKa = 11.84EE146 pKa = 4.34KK147 pKa = 10.45KK148 pKa = 10.42LKK150 pKa = 10.42LNRR153 pKa = 11.84DD154 pKa = 3.28VPDD157 pKa = 3.45VYY159 pKa = 11.46VRR161 pKa = 11.84IGPSGTGKK169 pKa = 8.13TRR171 pKa = 11.84WLDD174 pKa = 3.34DD175 pKa = 3.52TFGLDD180 pKa = 3.13GWTFAPDD187 pKa = 3.65NTGHH191 pKa = 6.86WFDD194 pKa = 5.0GCDD197 pKa = 3.54CDD199 pKa = 5.59VICFDD204 pKa = 4.2EE205 pKa = 4.91VEE207 pKa = 4.41AGAIPSLSTWKK218 pKa = 10.16ILCDD222 pKa = 3.58RR223 pKa = 11.84YY224 pKa = 10.82GKK226 pKa = 10.1RR227 pKa = 11.84VPVKK231 pKa = 10.6GGFIYY236 pKa = 9.77WKK238 pKa = 9.17PKK240 pKa = 10.38VIVFTSNQPISKK252 pKa = 8.42WWPNLTEE259 pKa = 4.19FDD261 pKa = 4.73LIAIEE266 pKa = 4.37RR267 pKa = 11.84RR268 pKa = 11.84CKK270 pKa = 10.4EE271 pKa = 3.84IVVVEE276 pKa = 4.1
MM1 pKa = 7.91PGYY4 pKa = 9.29QFRR7 pKa = 11.84SATFTQFNVEE17 pKa = 4.18KK18 pKa = 10.62APEE21 pKa = 4.47LGNLQYY27 pKa = 10.78IAFGLEE33 pKa = 4.31TCPSTQRR40 pKa = 11.84QHH42 pKa = 4.93WQGFAYY48 pKa = 10.21SYY50 pKa = 11.3TKK52 pKa = 8.21MTHH55 pKa = 5.52KK56 pKa = 10.1KK57 pKa = 8.59WRR59 pKa = 11.84EE60 pKa = 3.95TFPGAHH66 pKa = 5.97IEE68 pKa = 4.06EE69 pKa = 4.45MKK71 pKa = 11.19GNFRR75 pKa = 11.84QNEE78 pKa = 4.69VYY80 pKa = 10.2CSKK83 pKa = 10.7EE84 pKa = 3.51GTYY87 pKa = 10.1TEE89 pKa = 5.02FGIKK93 pKa = 10.02PMEE96 pKa = 4.42NGKK99 pKa = 9.73KK100 pKa = 9.94RR101 pKa = 11.84SLAEE105 pKa = 3.87MKK107 pKa = 10.23EE108 pKa = 3.86QLDD111 pKa = 3.71NGEE114 pKa = 4.48KK115 pKa = 9.72PVEE118 pKa = 4.13VATKK122 pKa = 9.07SDD124 pKa = 3.15KK125 pKa = 10.88MFGTFLQYY133 pKa = 10.62RR134 pKa = 11.84SGLNEE139 pKa = 3.42YY140 pKa = 9.88AQYY143 pKa = 10.58IRR145 pKa = 11.84EE146 pKa = 4.34KK147 pKa = 10.45KK148 pKa = 10.42LKK150 pKa = 10.42LNRR153 pKa = 11.84DD154 pKa = 3.28VPDD157 pKa = 3.45VYY159 pKa = 11.46VRR161 pKa = 11.84IGPSGTGKK169 pKa = 8.13TRR171 pKa = 11.84WLDD174 pKa = 3.34DD175 pKa = 3.52TFGLDD180 pKa = 3.13GWTFAPDD187 pKa = 3.65NTGHH191 pKa = 6.86WFDD194 pKa = 5.0GCDD197 pKa = 3.54CDD199 pKa = 5.59VICFDD204 pKa = 4.2EE205 pKa = 4.91VEE207 pKa = 4.41AGAIPSLSTWKK218 pKa = 10.16ILCDD222 pKa = 3.58RR223 pKa = 11.84YY224 pKa = 10.82GKK226 pKa = 10.1RR227 pKa = 11.84VPVKK231 pKa = 10.6GGFIYY236 pKa = 9.77WKK238 pKa = 9.17PKK240 pKa = 10.38VIVFTSNQPISKK252 pKa = 8.42WWPNLTEE259 pKa = 4.19FDD261 pKa = 4.73LIAIEE266 pKa = 4.37RR267 pKa = 11.84RR268 pKa = 11.84CKK270 pKa = 10.4EE271 pKa = 3.84IVVVEE276 pKa = 4.1
Molecular weight: 31.95 kDa
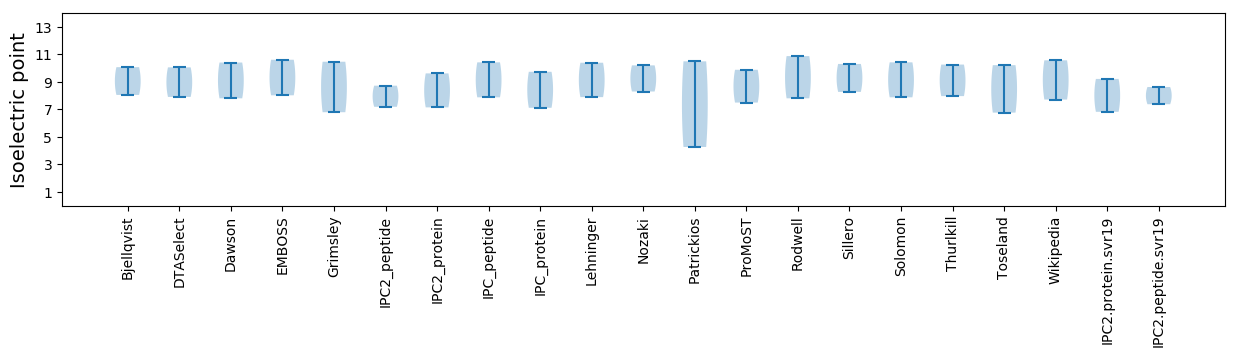
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A141MJB0|A0A141MJB0_9VIRU Uncharacterized protein OS=Ctenophore-associated circular virus 3 OX=1778560 PE=4 SV=1
MM1 pKa = 7.91PIGRR5 pKa = 11.84KK6 pKa = 8.77RR7 pKa = 11.84KK8 pKa = 9.38SYY10 pKa = 9.44ATAPYY15 pKa = 9.36IKK17 pKa = 9.8RR18 pKa = 11.84RR19 pKa = 11.84KK20 pKa = 6.98TPYY23 pKa = 8.92RR24 pKa = 11.84RR25 pKa = 11.84KK26 pKa = 10.2SFMSRR31 pKa = 11.84NVSRR35 pKa = 11.84IARR38 pKa = 11.84QVVMRR43 pKa = 11.84AAEE46 pKa = 4.23TKK48 pKa = 10.11TKK50 pKa = 10.15VINYY54 pKa = 7.65GKK56 pKa = 8.74TEE58 pKa = 4.51LYY60 pKa = 10.59HH61 pKa = 6.52NSLSVTHH68 pKa = 6.75KK69 pKa = 11.14LNFTAHH75 pKa = 6.06MPAQGDD81 pKa = 4.0GKK83 pKa = 10.27DD84 pKa = 3.68GRR86 pKa = 11.84SGDD89 pKa = 4.1SIIGKK94 pKa = 8.21GWKK97 pKa = 9.9LRR99 pKa = 11.84MTLANKK105 pKa = 9.97GDD107 pKa = 4.06RR108 pKa = 11.84PNVTYY113 pKa = 10.77RR114 pKa = 11.84LVVFAAKK121 pKa = 10.42AGVTPTYY128 pKa = 10.95SNTFVNVTGNGLLDD142 pKa = 4.99GINTDD147 pKa = 3.19RR148 pKa = 11.84CTILYY153 pKa = 8.57QKK155 pKa = 8.7WLKK158 pKa = 9.25PNKK161 pKa = 10.05GMVGTSEE168 pKa = 4.02TGVRR172 pKa = 11.84EE173 pKa = 4.07YY174 pKa = 11.57VSTKK178 pKa = 10.45SIWIKK183 pKa = 10.2RR184 pKa = 11.84EE185 pKa = 3.41KK186 pKa = 10.58VMKK189 pKa = 10.7FNGLEE194 pKa = 4.01VNDD197 pKa = 4.03RR198 pKa = 11.84DD199 pKa = 4.92VYY201 pKa = 11.13FCMLAYY207 pKa = 10.1DD208 pKa = 4.31AYY210 pKa = 10.22GTLTADD216 pKa = 3.86NIGYY220 pKa = 7.81MQMWGEE226 pKa = 4.45FKK228 pKa = 11.22YY229 pKa = 10.67KK230 pKa = 10.71DD231 pKa = 3.43PP232 pKa = 4.96
MM1 pKa = 7.91PIGRR5 pKa = 11.84KK6 pKa = 8.77RR7 pKa = 11.84KK8 pKa = 9.38SYY10 pKa = 9.44ATAPYY15 pKa = 9.36IKK17 pKa = 9.8RR18 pKa = 11.84RR19 pKa = 11.84KK20 pKa = 6.98TPYY23 pKa = 8.92RR24 pKa = 11.84RR25 pKa = 11.84KK26 pKa = 10.2SFMSRR31 pKa = 11.84NVSRR35 pKa = 11.84IARR38 pKa = 11.84QVVMRR43 pKa = 11.84AAEE46 pKa = 4.23TKK48 pKa = 10.11TKK50 pKa = 10.15VINYY54 pKa = 7.65GKK56 pKa = 8.74TEE58 pKa = 4.51LYY60 pKa = 10.59HH61 pKa = 6.52NSLSVTHH68 pKa = 6.75KK69 pKa = 11.14LNFTAHH75 pKa = 6.06MPAQGDD81 pKa = 4.0GKK83 pKa = 10.27DD84 pKa = 3.68GRR86 pKa = 11.84SGDD89 pKa = 4.1SIIGKK94 pKa = 8.21GWKK97 pKa = 9.9LRR99 pKa = 11.84MTLANKK105 pKa = 9.97GDD107 pKa = 4.06RR108 pKa = 11.84PNVTYY113 pKa = 10.77RR114 pKa = 11.84LVVFAAKK121 pKa = 10.42AGVTPTYY128 pKa = 10.95SNTFVNVTGNGLLDD142 pKa = 4.99GINTDD147 pKa = 3.19RR148 pKa = 11.84CTILYY153 pKa = 8.57QKK155 pKa = 8.7WLKK158 pKa = 9.25PNKK161 pKa = 10.05GMVGTSEE168 pKa = 4.02TGVRR172 pKa = 11.84EE173 pKa = 4.07YY174 pKa = 11.57VSTKK178 pKa = 10.45SIWIKK183 pKa = 10.2RR184 pKa = 11.84EE185 pKa = 3.41KK186 pKa = 10.58VMKK189 pKa = 10.7FNGLEE194 pKa = 4.01VNDD197 pKa = 4.03RR198 pKa = 11.84DD199 pKa = 4.92VYY201 pKa = 11.13FCMLAYY207 pKa = 10.1DD208 pKa = 4.31AYY210 pKa = 10.22GTLTADD216 pKa = 3.86NIGYY220 pKa = 7.81MQMWGEE226 pKa = 4.45FKK228 pKa = 11.22YY229 pKa = 10.67KK230 pKa = 10.71DD231 pKa = 3.43PP232 pKa = 4.96
Molecular weight: 26.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
508 |
232 |
276 |
254.0 |
29.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.118 ± 0.61 | 1.772 ± 0.605 |
4.921 ± 0.12 | 5.709 ± 1.791 |
4.724 ± 1.136 | 8.465 ± 0.104 |
1.378 ± 0.056 | 4.921 ± 0.12 |
9.252 ± 0.441 | 5.512 ± 0.061 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.15 ± 0.773 | 4.921 ± 0.741 |
4.331 ± 0.587 | 2.953 ± 0.818 |
6.299 ± 0.971 | 4.724 ± 0.298 |
7.677 ± 0.628 | 6.496 ± 0.553 |
2.559 ± 0.556 | 5.118 ± 0.61 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
