
Talaromyces atroroseus
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; Eurotiomycetes; Eurotiomycetidae; Eurotiales; Trichocomaceae; Talaromyces; Talaromyces sect. Trachyspermi
Average proteome isoelectric point is 6.28
Get precalculated fractions of proteins
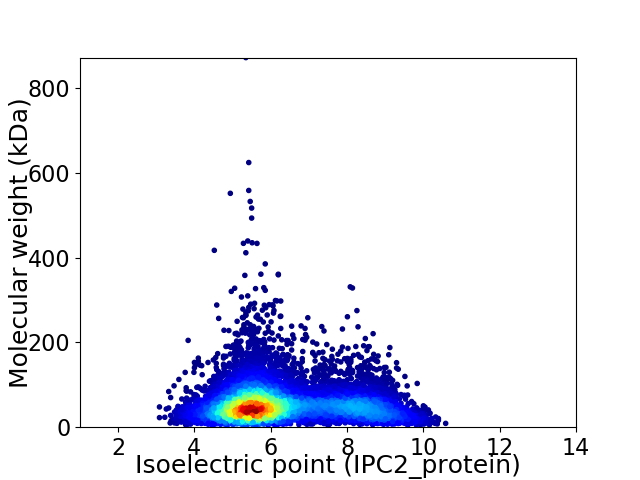
Virtual 2D-PAGE plot for 9520 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
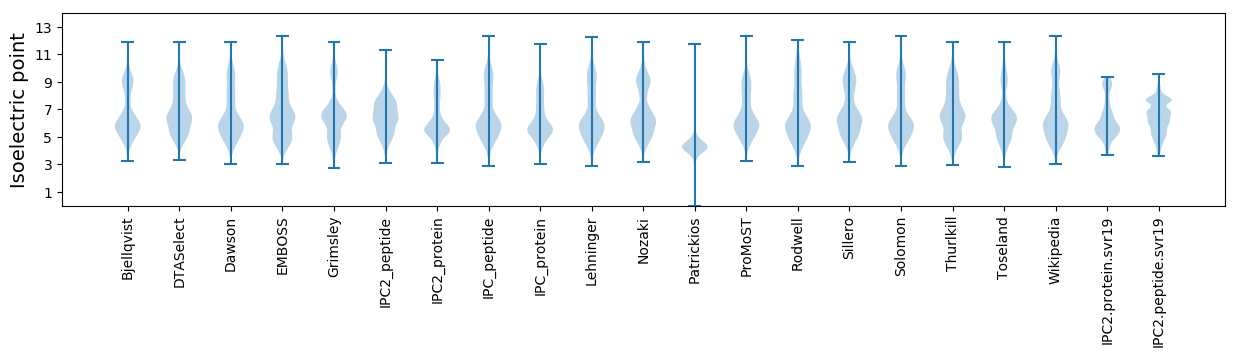
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A225ACX0|A0A225ACX0_9EURO Protein kinase gsk3 OS=Talaromyces atroroseus OX=1441469 GN=UA08_07899 PE=3 SV=1
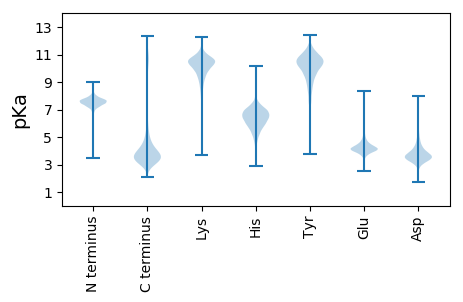
MM1 pKa = 7.57PSKK4 pKa = 9.88TGLALFGLAASALAEE19 pKa = 4.08PQVLHH24 pKa = 6.33MPLTRR29 pKa = 11.84NPNANRR35 pKa = 11.84LAKK38 pKa = 10.47RR39 pKa = 11.84NGDD42 pKa = 3.67ALVDD46 pKa = 3.52ITNDD50 pKa = 3.25LSEE53 pKa = 3.98GLYY56 pKa = 10.08YY57 pKa = 11.2VNASVGTPGQEE68 pKa = 3.56IQLTLDD74 pKa = 3.71TGSSDD79 pKa = 2.37IWFFGANSCDD89 pKa = 4.22EE90 pKa = 4.14NTTDD94 pKa = 5.52CFGGTYY100 pKa = 10.22NPSKK104 pKa = 10.43SSSVTVLNPKK114 pKa = 9.92GAFSIQYY121 pKa = 7.84GTAGSNVTGNYY132 pKa = 7.7ITDD135 pKa = 4.01DD136 pKa = 4.04FSVGAASVKK145 pKa = 10.15NLTMAYY151 pKa = 7.93ATYY154 pKa = 10.47AAYY157 pKa = 10.62VPTGVMGIGFDD168 pKa = 3.65TNEE171 pKa = 5.1AITDD175 pKa = 4.06EE176 pKa = 4.52YY177 pKa = 10.99GSPYY181 pKa = 10.28PNFVDD186 pKa = 3.78VLVSQNVTKK195 pKa = 10.51TKK197 pKa = 10.37AYY199 pKa = 10.47SLWLNDD205 pKa = 4.18LDD207 pKa = 4.75SSTGNILFGGYY218 pKa = 7.58DD219 pKa = 3.27TKK221 pKa = 11.17KK222 pKa = 10.77FSGEE226 pKa = 4.07LLTVDD231 pKa = 3.97IQPDD235 pKa = 3.58EE236 pKa = 4.09MSGEE240 pKa = 4.17ITSMTVAWTSLNVTANSKK258 pKa = 5.72TTQVTKK264 pKa = 10.96SDD266 pKa = 3.85FTSPALLDD274 pKa = 3.68SGTTITIVPDD284 pKa = 4.38DD285 pKa = 3.33IYY287 pKa = 11.47YY288 pKa = 11.23ALFEE292 pKa = 4.26YY293 pKa = 9.94FQAEE297 pKa = 4.1SDD299 pKa = 4.18GEE301 pKa = 3.81GDD303 pKa = 4.94AIVKK307 pKa = 10.42CSLLDD312 pKa = 3.79SSDD315 pKa = 3.18GTLDD319 pKa = 3.45FGFGGTGGPVVKK331 pKa = 10.81VPFSEE336 pKa = 4.4LALPATDD343 pKa = 4.83TNGNWLAFEE352 pKa = 5.07DD353 pKa = 4.84GEE355 pKa = 4.45LACVLGLEE363 pKa = 4.43GTDD366 pKa = 3.55DD367 pKa = 4.17GEE369 pKa = 4.37MPVIFGDD376 pKa = 3.54TFLRR380 pKa = 11.84SAYY383 pKa = 9.93VVYY386 pKa = 10.88DD387 pKa = 3.61LTNKK391 pKa = 10.0QISLAQTVFNATDD404 pKa = 3.53SNIVEE409 pKa = 4.53ISSSSPVASVVTGVTVTQTATNNPLGQATATAASSTPTDD448 pKa = 3.34SSGSSINSIGVLSSSTSTATSAASTSSHH476 pKa = 5.89GSAASIPAPVPGLMTSLLVAGASMMFGSVFFMLHH510 pKa = 5.43
MM1 pKa = 7.57PSKK4 pKa = 9.88TGLALFGLAASALAEE19 pKa = 4.08PQVLHH24 pKa = 6.33MPLTRR29 pKa = 11.84NPNANRR35 pKa = 11.84LAKK38 pKa = 10.47RR39 pKa = 11.84NGDD42 pKa = 3.67ALVDD46 pKa = 3.52ITNDD50 pKa = 3.25LSEE53 pKa = 3.98GLYY56 pKa = 10.08YY57 pKa = 11.2VNASVGTPGQEE68 pKa = 3.56IQLTLDD74 pKa = 3.71TGSSDD79 pKa = 2.37IWFFGANSCDD89 pKa = 4.22EE90 pKa = 4.14NTTDD94 pKa = 5.52CFGGTYY100 pKa = 10.22NPSKK104 pKa = 10.43SSSVTVLNPKK114 pKa = 9.92GAFSIQYY121 pKa = 7.84GTAGSNVTGNYY132 pKa = 7.7ITDD135 pKa = 4.01DD136 pKa = 4.04FSVGAASVKK145 pKa = 10.15NLTMAYY151 pKa = 7.93ATYY154 pKa = 10.47AAYY157 pKa = 10.62VPTGVMGIGFDD168 pKa = 3.65TNEE171 pKa = 5.1AITDD175 pKa = 4.06EE176 pKa = 4.52YY177 pKa = 10.99GSPYY181 pKa = 10.28PNFVDD186 pKa = 3.78VLVSQNVTKK195 pKa = 10.51TKK197 pKa = 10.37AYY199 pKa = 10.47SLWLNDD205 pKa = 4.18LDD207 pKa = 4.75SSTGNILFGGYY218 pKa = 7.58DD219 pKa = 3.27TKK221 pKa = 11.17KK222 pKa = 10.77FSGEE226 pKa = 4.07LLTVDD231 pKa = 3.97IQPDD235 pKa = 3.58EE236 pKa = 4.09MSGEE240 pKa = 4.17ITSMTVAWTSLNVTANSKK258 pKa = 5.72TTQVTKK264 pKa = 10.96SDD266 pKa = 3.85FTSPALLDD274 pKa = 3.68SGTTITIVPDD284 pKa = 4.38DD285 pKa = 3.33IYY287 pKa = 11.47YY288 pKa = 11.23ALFEE292 pKa = 4.26YY293 pKa = 9.94FQAEE297 pKa = 4.1SDD299 pKa = 4.18GEE301 pKa = 3.81GDD303 pKa = 4.94AIVKK307 pKa = 10.42CSLLDD312 pKa = 3.79SSDD315 pKa = 3.18GTLDD319 pKa = 3.45FGFGGTGGPVVKK331 pKa = 10.81VPFSEE336 pKa = 4.4LALPATDD343 pKa = 4.83TNGNWLAFEE352 pKa = 5.07DD353 pKa = 4.84GEE355 pKa = 4.45LACVLGLEE363 pKa = 4.43GTDD366 pKa = 3.55DD367 pKa = 4.17GEE369 pKa = 4.37MPVIFGDD376 pKa = 3.54TFLRR380 pKa = 11.84SAYY383 pKa = 9.93VVYY386 pKa = 10.88DD387 pKa = 3.61LTNKK391 pKa = 10.0QISLAQTVFNATDD404 pKa = 3.53SNIVEE409 pKa = 4.53ISSSSPVASVVTGVTVTQTATNNPLGQATATAASSTPTDD448 pKa = 3.34SSGSSINSIGVLSSSTSTATSAASTSSHH476 pKa = 5.89GSAASIPAPVPGLMTSLLVAGASMMFGSVFFMLHH510 pKa = 5.43
Molecular weight: 53.02 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Q5Q693|A0A1Q5Q693_9EURO Uncharacterized protein OS=Talaromyces atroroseus OX=1441469 GN=UA08_09365 PE=3 SV=1
MM1 pKa = 8.0AIPRR5 pKa = 11.84TPRR8 pKa = 11.84TMAKK12 pKa = 10.16HH13 pKa = 5.7PMPPFLRR20 pKa = 11.84QEE22 pKa = 4.12EE23 pKa = 4.34EE24 pKa = 4.09KK25 pKa = 10.39MRR27 pKa = 11.84MEE29 pKa = 4.81SPRR32 pKa = 11.84FACPPKK38 pKa = 9.29NTAIKK43 pKa = 7.75TTIKK47 pKa = 7.01TTLMKK52 pKa = 10.24PSLISVVHH60 pKa = 6.74GKK62 pKa = 8.98RR63 pKa = 11.84QPWRR67 pKa = 11.84HH68 pKa = 4.06MWIPRR73 pKa = 11.84RR74 pKa = 11.84PRR76 pKa = 11.84THH78 pKa = 6.5CWKK81 pKa = 9.96SACPGPVQRR90 pKa = 11.84LLTSVLTRR98 pKa = 11.84PTGQRR103 pKa = 11.84SPRR106 pKa = 11.84NSNYY110 pKa = 9.82LRR112 pKa = 11.84LILKK116 pKa = 8.93CQFWANGARR125 pKa = 11.84KK126 pKa = 9.58HH127 pKa = 5.79ILNVRR132 pKa = 11.84NGKK135 pKa = 10.25RR136 pKa = 11.84IDD138 pKa = 3.67SQEE141 pKa = 3.8VLGDD145 pKa = 3.62STRR148 pKa = 11.84ATAATKK154 pKa = 9.29RR155 pKa = 11.84TAPHH159 pKa = 6.48HH160 pKa = 7.05DD161 pKa = 3.97SYY163 pKa = 11.56NSPVGRR169 pKa = 11.84LIGEE173 pKa = 4.1GRR175 pKa = 11.84VRR177 pKa = 11.84SPGLFAIRR185 pKa = 11.84NDD187 pKa = 3.73HH188 pKa = 7.44DD189 pKa = 4.1GGGYY193 pKa = 8.36GTDD196 pKa = 3.67DD197 pKa = 4.0SGSASASRR205 pKa = 11.84GSIFSRR211 pKa = 11.84QSEE214 pKa = 4.02KK215 pKa = 10.46RR216 pKa = 11.84YY217 pKa = 10.1SGSVTSGEE225 pKa = 4.31GPSPNSLVDD234 pKa = 3.82PGAEE238 pKa = 3.41QSGFSRR244 pKa = 11.84AAGGQSRR251 pKa = 11.84VPAEE255 pKa = 4.31SPSVASVSRR264 pKa = 11.84QLGSEE269 pKa = 4.08IEE271 pKa = 4.32DD272 pKa = 3.73SEE274 pKa = 4.76SASDD278 pKa = 3.53TDD280 pKa = 5.2DD281 pKa = 4.74ADD283 pKa = 3.49HH284 pKa = 7.16HH285 pKa = 7.34SGDD288 pKa = 3.54GRR290 pKa = 11.84WQYY293 pKa = 11.54RR294 pKa = 11.84CILKK298 pKa = 10.32RR299 pKa = 11.84RR300 pKa = 11.84LDD302 pKa = 3.48SSGRR306 pKa = 11.84RR307 pKa = 11.84MALVRR312 pKa = 11.84WEE314 pKa = 4.36DD315 pKa = 3.07TWEE318 pKa = 4.37SEE320 pKa = 4.99DD321 pKa = 3.56EE322 pKa = 4.17LGGVKK327 pKa = 10.05RR328 pKa = 11.84ALRR331 pKa = 11.84QYY333 pKa = 11.34ARR335 pKa = 11.84ARR337 pKa = 11.84QAKK340 pKa = 8.37QVPATEE346 pKa = 4.46TCSKK350 pKa = 10.3RR351 pKa = 11.84RR352 pKa = 11.84GRR354 pKa = 11.84KK355 pKa = 8.8RR356 pKa = 11.84KK357 pKa = 9.9CPSS360 pKa = 2.78
MM1 pKa = 8.0AIPRR5 pKa = 11.84TPRR8 pKa = 11.84TMAKK12 pKa = 10.16HH13 pKa = 5.7PMPPFLRR20 pKa = 11.84QEE22 pKa = 4.12EE23 pKa = 4.34EE24 pKa = 4.09KK25 pKa = 10.39MRR27 pKa = 11.84MEE29 pKa = 4.81SPRR32 pKa = 11.84FACPPKK38 pKa = 9.29NTAIKK43 pKa = 7.75TTIKK47 pKa = 7.01TTLMKK52 pKa = 10.24PSLISVVHH60 pKa = 6.74GKK62 pKa = 8.98RR63 pKa = 11.84QPWRR67 pKa = 11.84HH68 pKa = 4.06MWIPRR73 pKa = 11.84RR74 pKa = 11.84PRR76 pKa = 11.84THH78 pKa = 6.5CWKK81 pKa = 9.96SACPGPVQRR90 pKa = 11.84LLTSVLTRR98 pKa = 11.84PTGQRR103 pKa = 11.84SPRR106 pKa = 11.84NSNYY110 pKa = 9.82LRR112 pKa = 11.84LILKK116 pKa = 8.93CQFWANGARR125 pKa = 11.84KK126 pKa = 9.58HH127 pKa = 5.79ILNVRR132 pKa = 11.84NGKK135 pKa = 10.25RR136 pKa = 11.84IDD138 pKa = 3.67SQEE141 pKa = 3.8VLGDD145 pKa = 3.62STRR148 pKa = 11.84ATAATKK154 pKa = 9.29RR155 pKa = 11.84TAPHH159 pKa = 6.48HH160 pKa = 7.05DD161 pKa = 3.97SYY163 pKa = 11.56NSPVGRR169 pKa = 11.84LIGEE173 pKa = 4.1GRR175 pKa = 11.84VRR177 pKa = 11.84SPGLFAIRR185 pKa = 11.84NDD187 pKa = 3.73HH188 pKa = 7.44DD189 pKa = 4.1GGGYY193 pKa = 8.36GTDD196 pKa = 3.67DD197 pKa = 4.0SGSASASRR205 pKa = 11.84GSIFSRR211 pKa = 11.84QSEE214 pKa = 4.02KK215 pKa = 10.46RR216 pKa = 11.84YY217 pKa = 10.1SGSVTSGEE225 pKa = 4.31GPSPNSLVDD234 pKa = 3.82PGAEE238 pKa = 3.41QSGFSRR244 pKa = 11.84AAGGQSRR251 pKa = 11.84VPAEE255 pKa = 4.31SPSVASVSRR264 pKa = 11.84QLGSEE269 pKa = 4.08IEE271 pKa = 4.32DD272 pKa = 3.73SEE274 pKa = 4.76SASDD278 pKa = 3.53TDD280 pKa = 5.2DD281 pKa = 4.74ADD283 pKa = 3.49HH284 pKa = 7.16HH285 pKa = 7.34SGDD288 pKa = 3.54GRR290 pKa = 11.84WQYY293 pKa = 11.54RR294 pKa = 11.84CILKK298 pKa = 10.32RR299 pKa = 11.84RR300 pKa = 11.84LDD302 pKa = 3.48SSGRR306 pKa = 11.84RR307 pKa = 11.84MALVRR312 pKa = 11.84WEE314 pKa = 4.36DD315 pKa = 3.07TWEE318 pKa = 4.37SEE320 pKa = 4.99DD321 pKa = 3.56EE322 pKa = 4.17LGGVKK327 pKa = 10.05RR328 pKa = 11.84ALRR331 pKa = 11.84QYY333 pKa = 11.34ARR335 pKa = 11.84ARR337 pKa = 11.84QAKK340 pKa = 8.37QVPATEE346 pKa = 4.46TCSKK350 pKa = 10.3RR351 pKa = 11.84RR352 pKa = 11.84GRR354 pKa = 11.84KK355 pKa = 8.8RR356 pKa = 11.84KK357 pKa = 9.9CPSS360 pKa = 2.78
Molecular weight: 40.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5014686 |
49 |
7850 |
526.8 |
58.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.365 ± 0.017 | 1.16 ± 0.009 |
5.705 ± 0.016 | 6.164 ± 0.024 |
3.797 ± 0.012 | 6.559 ± 0.021 |
2.401 ± 0.012 | 5.277 ± 0.016 |
4.7 ± 0.023 | 9.046 ± 0.028 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.094 ± 0.009 | 3.938 ± 0.012 |
5.666 ± 0.024 | 4.081 ± 0.016 |
5.939 ± 0.021 | 8.632 ± 0.029 |
6.03 ± 0.02 | 6.117 ± 0.017 |
1.427 ± 0.009 | 2.904 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
