
Codonopsis vein clearing virus
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Ortervirales; Caulimoviridae; Badnavirus
Average proteome isoelectric point is 6.74
Get precalculated fractions of proteins
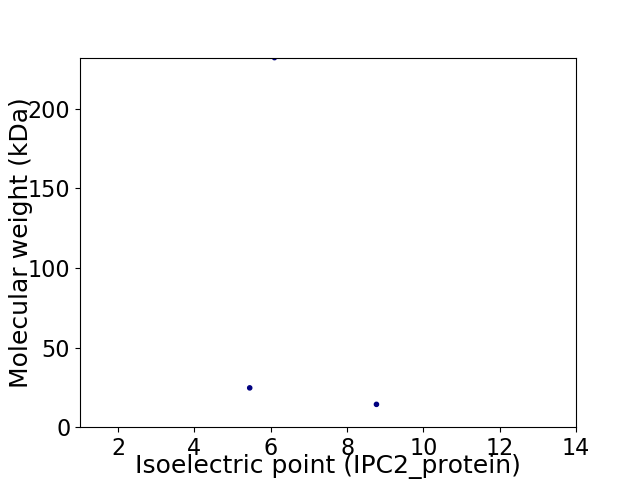
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A411AUG2|A0A411AUG2_9VIRU ORF2 OS=Codonopsis vein clearing virus OX=2510980 PE=4 SV=1
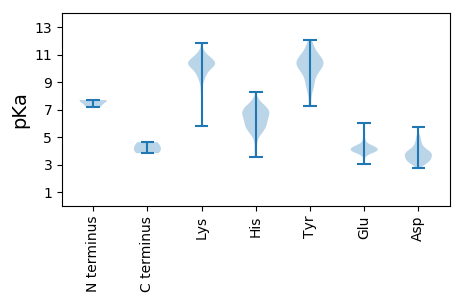
MM1 pKa = 7.66RR2 pKa = 11.84AALEE6 pKa = 4.09CKK8 pKa = 9.57EE9 pKa = 4.18FEE11 pKa = 4.37EE12 pKa = 5.23EE13 pKa = 4.14VEE15 pKa = 3.84KK16 pKa = 11.02WEE18 pKa = 4.14IPEE21 pKa = 4.21SSTLAGFKK29 pKa = 10.32EE30 pKa = 4.2LEE32 pKa = 4.02QYY34 pKa = 10.67PNLRR38 pKa = 11.84KK39 pKa = 9.82NVVYY43 pKa = 7.33PTCKK47 pKa = 10.09LPCFHH52 pKa = 7.25FSALADD58 pKa = 4.06PDD60 pKa = 3.38AHH62 pKa = 6.06ATFTKK67 pKa = 10.59RR68 pKa = 11.84EE69 pKa = 3.97DD70 pKa = 3.27RR71 pKa = 11.84VPFVVNTLFDD81 pKa = 3.88LAIIHH86 pKa = 5.69QVNLSYY92 pKa = 10.87IHH94 pKa = 6.7TRR96 pKa = 11.84LQLLSRR102 pKa = 11.84YY103 pKa = 7.23QARR106 pKa = 11.84KK107 pKa = 5.8TTGAPPLATIPEE119 pKa = 4.36DD120 pKa = 3.82RR121 pKa = 11.84PLPDD125 pKa = 4.19LSPVVEE131 pKa = 4.38AVTDD135 pKa = 3.82SVTGLKK141 pKa = 10.02SQIVSLKK148 pKa = 10.45ADD150 pKa = 3.39LRR152 pKa = 11.84EE153 pKa = 4.06IKK155 pKa = 10.48AGQTSIKK162 pKa = 10.2LALSDD167 pKa = 3.54LRR169 pKa = 11.84EE170 pKa = 4.18VVTALTEE177 pKa = 4.09RR178 pKa = 11.84EE179 pKa = 4.43LDD181 pKa = 3.78PKK183 pKa = 10.57PIEE186 pKa = 4.51ADD188 pKa = 3.34TAKK191 pKa = 10.54VAEE194 pKa = 4.02QLKK197 pKa = 10.33VSVKK201 pKa = 10.2EE202 pKa = 3.99IQASLKK208 pKa = 10.3DD209 pKa = 3.77LKK211 pKa = 11.13DD212 pKa = 3.55FAKK215 pKa = 10.77SLVPEE220 pKa = 4.34AA221 pKa = 4.62
MM1 pKa = 7.66RR2 pKa = 11.84AALEE6 pKa = 4.09CKK8 pKa = 9.57EE9 pKa = 4.18FEE11 pKa = 4.37EE12 pKa = 5.23EE13 pKa = 4.14VEE15 pKa = 3.84KK16 pKa = 11.02WEE18 pKa = 4.14IPEE21 pKa = 4.21SSTLAGFKK29 pKa = 10.32EE30 pKa = 4.2LEE32 pKa = 4.02QYY34 pKa = 10.67PNLRR38 pKa = 11.84KK39 pKa = 9.82NVVYY43 pKa = 7.33PTCKK47 pKa = 10.09LPCFHH52 pKa = 7.25FSALADD58 pKa = 4.06PDD60 pKa = 3.38AHH62 pKa = 6.06ATFTKK67 pKa = 10.59RR68 pKa = 11.84EE69 pKa = 3.97DD70 pKa = 3.27RR71 pKa = 11.84VPFVVNTLFDD81 pKa = 3.88LAIIHH86 pKa = 5.69QVNLSYY92 pKa = 10.87IHH94 pKa = 6.7TRR96 pKa = 11.84LQLLSRR102 pKa = 11.84YY103 pKa = 7.23QARR106 pKa = 11.84KK107 pKa = 5.8TTGAPPLATIPEE119 pKa = 4.36DD120 pKa = 3.82RR121 pKa = 11.84PLPDD125 pKa = 4.19LSPVVEE131 pKa = 4.38AVTDD135 pKa = 3.82SVTGLKK141 pKa = 10.02SQIVSLKK148 pKa = 10.45ADD150 pKa = 3.39LRR152 pKa = 11.84EE153 pKa = 4.06IKK155 pKa = 10.48AGQTSIKK162 pKa = 10.2LALSDD167 pKa = 3.54LRR169 pKa = 11.84EE170 pKa = 4.18VVTALTEE177 pKa = 4.09RR178 pKa = 11.84EE179 pKa = 4.43LDD181 pKa = 3.78PKK183 pKa = 10.57PIEE186 pKa = 4.51ADD188 pKa = 3.34TAKK191 pKa = 10.54VAEE194 pKa = 4.02QLKK197 pKa = 10.33VSVKK201 pKa = 10.2EE202 pKa = 3.99IQASLKK208 pKa = 10.3DD209 pKa = 3.77LKK211 pKa = 11.13DD212 pKa = 3.55FAKK215 pKa = 10.77SLVPEE220 pKa = 4.34AA221 pKa = 4.62
Molecular weight: 24.69 kDa
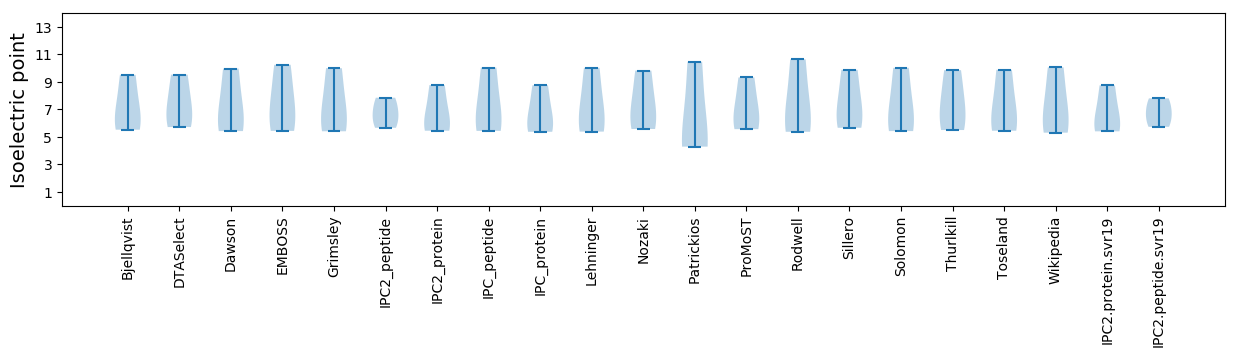
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A411AUG2|A0A411AUG2_9VIRU ORF2 OS=Codonopsis vein clearing virus OX=2510980 PE=4 SV=1
MM1 pKa = 7.69SSWQLAASTDD11 pKa = 3.81EE12 pKa = 3.84YY13 pKa = 11.28RR14 pKa = 11.84KK15 pKa = 10.31ALNATGSLTKK25 pKa = 10.46DD26 pKa = 3.15NRR28 pKa = 11.84PVGFVEE34 pKa = 3.84PHH36 pKa = 6.49LFQPNLGDD44 pKa = 4.12TNIQRR49 pKa = 11.84QNNTLIHH56 pKa = 6.6LLIQTLEE63 pKa = 4.07EE64 pKa = 4.2VKK66 pKa = 10.68ALRR69 pKa = 11.84DD70 pKa = 3.3QVQILRR76 pKa = 11.84DD77 pKa = 3.74QVDD80 pKa = 3.69SLSKK84 pKa = 10.73GKK86 pKa = 10.44APEE89 pKa = 3.91VQLPKK94 pKa = 10.93DD95 pKa = 3.95VVDD98 pKa = 4.92KK99 pKa = 11.16LAAQLTTTHH108 pKa = 5.54VTPKK112 pKa = 10.63GKK114 pKa = 9.94FRR116 pKa = 11.84GKK118 pKa = 10.33GKK120 pKa = 10.19EE121 pKa = 3.79GQFRR125 pKa = 11.84VWSS128 pKa = 3.83
MM1 pKa = 7.69SSWQLAASTDD11 pKa = 3.81EE12 pKa = 3.84YY13 pKa = 11.28RR14 pKa = 11.84KK15 pKa = 10.31ALNATGSLTKK25 pKa = 10.46DD26 pKa = 3.15NRR28 pKa = 11.84PVGFVEE34 pKa = 3.84PHH36 pKa = 6.49LFQPNLGDD44 pKa = 4.12TNIQRR49 pKa = 11.84QNNTLIHH56 pKa = 6.6LLIQTLEE63 pKa = 4.07EE64 pKa = 4.2VKK66 pKa = 10.68ALRR69 pKa = 11.84DD70 pKa = 3.3QVQILRR76 pKa = 11.84DD77 pKa = 3.74QVDD80 pKa = 3.69SLSKK84 pKa = 10.73GKK86 pKa = 10.44APEE89 pKa = 3.91VQLPKK94 pKa = 10.93DD95 pKa = 3.95VVDD98 pKa = 4.92KK99 pKa = 11.16LAAQLTTTHH108 pKa = 5.54VTPKK112 pKa = 10.63GKK114 pKa = 9.94FRR116 pKa = 11.84GKK118 pKa = 10.33GKK120 pKa = 10.19EE121 pKa = 3.79GQFRR125 pKa = 11.84VWSS128 pKa = 3.83
Molecular weight: 14.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2404 |
128 |
2055 |
801.3 |
90.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.947 ± 1.06 | 1.581 ± 0.394 |
5.075 ± 0.467 | 8.403 ± 0.778 |
3.245 ± 0.131 | 5.865 ± 1.465 |
2.163 ± 0.121 | 5.74 ± 0.864 |
7.488 ± 0.408 | 8.444 ± 1.805 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.121 ± 0.781 | 3.785 ± 0.678 |
5.699 ± 0.379 | 4.201 ± 0.928 |
5.408 ± 0.154 | 6.614 ± 0.312 |
6.406 ± 0.392 | 6.073 ± 0.982 |
1.331 ± 0.308 | 3.411 ± 0.982 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
