
Tursiops truncatus papillomavirus 5
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Omikronpapillomavirus; Omikronpapillomavirus 1
Average proteome isoelectric point is 6.28
Get precalculated fractions of proteins
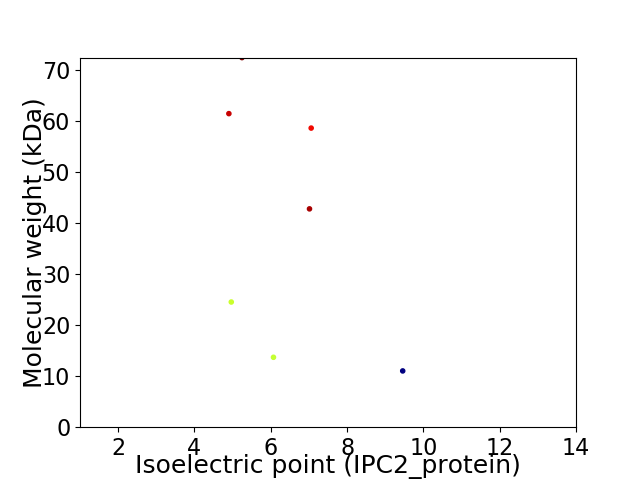
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|H6UYP4|H6UYP4_PSPV Major capsid protein L1 OS=Tursiops truncatus papillomavirus 5 OX=1144381 GN=L1 PE=3 SV=1
MM1 pKa = 6.86VRR3 pKa = 11.84ANRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84AAEE12 pKa = 3.55KK13 pKa = 10.47DD14 pKa = 3.39LYY16 pKa = 10.53AGCRR20 pKa = 11.84AGQDD24 pKa = 3.44CLDD27 pKa = 4.88DD28 pKa = 3.58IKK30 pKa = 11.35QKK32 pKa = 10.93FEE34 pKa = 4.22QNTVADD40 pKa = 6.13RR41 pKa = 11.84ILKK44 pKa = 8.67WLSSFIYY51 pKa = 10.15LGNLGISTGRR61 pKa = 11.84GSGGGSGYY69 pKa = 10.7VPIGSGGRR77 pKa = 11.84TVRR80 pKa = 11.84PAMGGQPSRR89 pKa = 11.84PNIVVEE95 pKa = 4.13NVGPAEE101 pKa = 4.18VPVGGTIDD109 pKa = 3.5ASAPSVITPSEE120 pKa = 4.09STVVVEE126 pKa = 5.17GATTSHH132 pKa = 6.48EE133 pKa = 4.56EE134 pKa = 3.86IPLVPLHH141 pKa = 7.05PDD143 pKa = 3.18VGAGGPDD150 pKa = 3.33PSLPVAPPVDD160 pKa = 3.45VDD162 pKa = 4.13GPAVLDD168 pKa = 3.6VSLDD172 pKa = 3.54VTSTYY177 pKa = 10.62PHH179 pKa = 7.41DD180 pKa = 4.27PSIVNPGSEE189 pKa = 4.31TLSTQLPSQFEE200 pKa = 4.37RR201 pKa = 11.84PGTSPLFPTVSLQPLDD217 pKa = 3.97VSLLPGEE224 pKa = 4.63TSFPPHH230 pKa = 5.28TVINLSGTFEE240 pKa = 4.34EE241 pKa = 5.29IEE243 pKa = 4.39LDD245 pKa = 3.79VLGNTLDD252 pKa = 3.98SEE254 pKa = 4.35PLYY257 pKa = 10.25PQTSTPKK264 pKa = 10.3SRR266 pKa = 11.84LDD268 pKa = 3.33TALTSVRR275 pKa = 11.84RR276 pKa = 11.84AYY278 pKa = 10.56NRR280 pKa = 11.84RR281 pKa = 11.84TDD283 pKa = 3.42VLRR286 pKa = 11.84KK287 pKa = 8.94YY288 pKa = 8.06YY289 pKa = 10.39HH290 pKa = 7.44RR291 pKa = 11.84LTQQIQVKK299 pKa = 10.01KK300 pKa = 10.38PEE302 pKa = 4.08FLQGPSQLVSYY313 pKa = 8.88EE314 pKa = 4.04FSNQAFDD321 pKa = 4.15PDD323 pKa = 3.53TTLFYY328 pKa = 11.03SQSTEE333 pKa = 4.07NVVTAPDD340 pKa = 3.78TDD342 pKa = 4.0FQDD345 pKa = 2.64IGTLHH350 pKa = 6.68RR351 pKa = 11.84PVYY354 pKa = 10.23SIEE357 pKa = 4.18GEE359 pKa = 4.11HH360 pKa = 6.41IRR362 pKa = 11.84VSRR365 pKa = 11.84FGEE368 pKa = 3.95RR369 pKa = 11.84EE370 pKa = 3.98TIRR373 pKa = 11.84TRR375 pKa = 11.84SGATIGAKK383 pKa = 8.27VHH385 pKa = 6.94FYY387 pKa = 10.76TDD389 pKa = 3.87LSSISHH395 pKa = 6.97LADD398 pKa = 3.08TFTSGTTLDD407 pKa = 4.61PDD409 pKa = 3.38IADD412 pKa = 4.26PGIEE416 pKa = 3.64LHH418 pKa = 6.89LFGEE422 pKa = 4.74STEE425 pKa = 4.32DD426 pKa = 3.06TSIAHH431 pKa = 5.66GQGGGIQFQNGEE443 pKa = 4.39MHH445 pKa = 6.83TEE447 pKa = 3.89TQFIDD452 pKa = 3.6VSNGSLHH459 pKa = 6.91SEE461 pKa = 4.42YY462 pKa = 10.47SDD464 pKa = 3.35SMLMDD469 pKa = 4.25TYY471 pKa = 11.59SEE473 pKa = 4.47TFSNAHH479 pKa = 6.37LALLNSDD486 pKa = 3.92SSAQLMSIPEE496 pKa = 4.05LARR499 pKa = 11.84PVRR502 pKa = 11.84EE503 pKa = 4.03LAEE506 pKa = 4.39STGGLSVLYY515 pKa = 8.97PVNGSVDD522 pKa = 4.18LSTSSPSFIDD532 pKa = 3.58NKK534 pKa = 9.95PIPPFIFFLFSHH546 pKa = 7.08GDD548 pKa = 3.3PSFFLHH554 pKa = 7.12PSLLRR559 pKa = 11.84KK560 pKa = 8.78KK561 pKa = 10.06RR562 pKa = 11.84KK563 pKa = 8.94RR564 pKa = 11.84VFYY567 pKa = 10.92
MM1 pKa = 6.86VRR3 pKa = 11.84ANRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84AAEE12 pKa = 3.55KK13 pKa = 10.47DD14 pKa = 3.39LYY16 pKa = 10.53AGCRR20 pKa = 11.84AGQDD24 pKa = 3.44CLDD27 pKa = 4.88DD28 pKa = 3.58IKK30 pKa = 11.35QKK32 pKa = 10.93FEE34 pKa = 4.22QNTVADD40 pKa = 6.13RR41 pKa = 11.84ILKK44 pKa = 8.67WLSSFIYY51 pKa = 10.15LGNLGISTGRR61 pKa = 11.84GSGGGSGYY69 pKa = 10.7VPIGSGGRR77 pKa = 11.84TVRR80 pKa = 11.84PAMGGQPSRR89 pKa = 11.84PNIVVEE95 pKa = 4.13NVGPAEE101 pKa = 4.18VPVGGTIDD109 pKa = 3.5ASAPSVITPSEE120 pKa = 4.09STVVVEE126 pKa = 5.17GATTSHH132 pKa = 6.48EE133 pKa = 4.56EE134 pKa = 3.86IPLVPLHH141 pKa = 7.05PDD143 pKa = 3.18VGAGGPDD150 pKa = 3.33PSLPVAPPVDD160 pKa = 3.45VDD162 pKa = 4.13GPAVLDD168 pKa = 3.6VSLDD172 pKa = 3.54VTSTYY177 pKa = 10.62PHH179 pKa = 7.41DD180 pKa = 4.27PSIVNPGSEE189 pKa = 4.31TLSTQLPSQFEE200 pKa = 4.37RR201 pKa = 11.84PGTSPLFPTVSLQPLDD217 pKa = 3.97VSLLPGEE224 pKa = 4.63TSFPPHH230 pKa = 5.28TVINLSGTFEE240 pKa = 4.34EE241 pKa = 5.29IEE243 pKa = 4.39LDD245 pKa = 3.79VLGNTLDD252 pKa = 3.98SEE254 pKa = 4.35PLYY257 pKa = 10.25PQTSTPKK264 pKa = 10.3SRR266 pKa = 11.84LDD268 pKa = 3.33TALTSVRR275 pKa = 11.84RR276 pKa = 11.84AYY278 pKa = 10.56NRR280 pKa = 11.84RR281 pKa = 11.84TDD283 pKa = 3.42VLRR286 pKa = 11.84KK287 pKa = 8.94YY288 pKa = 8.06YY289 pKa = 10.39HH290 pKa = 7.44RR291 pKa = 11.84LTQQIQVKK299 pKa = 10.01KK300 pKa = 10.38PEE302 pKa = 4.08FLQGPSQLVSYY313 pKa = 8.88EE314 pKa = 4.04FSNQAFDD321 pKa = 4.15PDD323 pKa = 3.53TTLFYY328 pKa = 11.03SQSTEE333 pKa = 4.07NVVTAPDD340 pKa = 3.78TDD342 pKa = 4.0FQDD345 pKa = 2.64IGTLHH350 pKa = 6.68RR351 pKa = 11.84PVYY354 pKa = 10.23SIEE357 pKa = 4.18GEE359 pKa = 4.11HH360 pKa = 6.41IRR362 pKa = 11.84VSRR365 pKa = 11.84FGEE368 pKa = 3.95RR369 pKa = 11.84EE370 pKa = 3.98TIRR373 pKa = 11.84TRR375 pKa = 11.84SGATIGAKK383 pKa = 8.27VHH385 pKa = 6.94FYY387 pKa = 10.76TDD389 pKa = 3.87LSSISHH395 pKa = 6.97LADD398 pKa = 3.08TFTSGTTLDD407 pKa = 4.61PDD409 pKa = 3.38IADD412 pKa = 4.26PGIEE416 pKa = 3.64LHH418 pKa = 6.89LFGEE422 pKa = 4.74STEE425 pKa = 4.32DD426 pKa = 3.06TSIAHH431 pKa = 5.66GQGGGIQFQNGEE443 pKa = 4.39MHH445 pKa = 6.83TEE447 pKa = 3.89TQFIDD452 pKa = 3.6VSNGSLHH459 pKa = 6.91SEE461 pKa = 4.42YY462 pKa = 10.47SDD464 pKa = 3.35SMLMDD469 pKa = 4.25TYY471 pKa = 11.59SEE473 pKa = 4.47TFSNAHH479 pKa = 6.37LALLNSDD486 pKa = 3.92SSAQLMSIPEE496 pKa = 4.05LARR499 pKa = 11.84PVRR502 pKa = 11.84EE503 pKa = 4.03LAEE506 pKa = 4.39STGGLSVLYY515 pKa = 8.97PVNGSVDD522 pKa = 4.18LSTSSPSFIDD532 pKa = 3.58NKK534 pKa = 9.95PIPPFIFFLFSHH546 pKa = 7.08GDD548 pKa = 3.3PSFFLHH554 pKa = 7.12PSLLRR559 pKa = 11.84KK560 pKa = 8.78KK561 pKa = 10.06RR562 pKa = 11.84KK563 pKa = 8.94RR564 pKa = 11.84VFYY567 pKa = 10.92
Molecular weight: 61.46 kDa
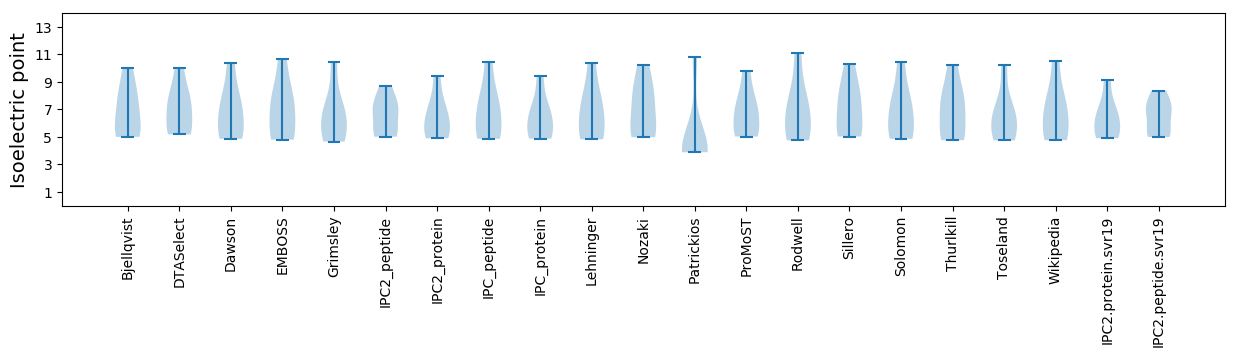
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|H6UYP5|H6UYP5_PSPV L3 OS=Tursiops truncatus papillomavirus 5 OX=1144381 GN=L3 PE=4 SV=1
MM1 pKa = 7.29GNSLIGHH8 pKa = 6.44IGFRR12 pKa = 11.84LPRR15 pKa = 11.84EE16 pKa = 4.05KK17 pKa = 10.63IMGYY21 pKa = 8.9VGEE24 pKa = 4.63MNFLSQWLIILEE36 pKa = 4.14TLILQLVLKK45 pKa = 6.77TQKK48 pKa = 10.91NNWTKK53 pKa = 10.67VPFKK57 pKa = 11.07QMLSKK62 pKa = 9.48FTQGIWRR69 pKa = 11.84NMSLISSFSSAKK81 pKa = 9.4CHH83 pKa = 6.24YY84 pKa = 10.29SLKK87 pKa = 10.44CWLKK91 pKa = 10.96YY92 pKa = 8.82ITT94 pKa = 4.65
MM1 pKa = 7.29GNSLIGHH8 pKa = 6.44IGFRR12 pKa = 11.84LPRR15 pKa = 11.84EE16 pKa = 4.05KK17 pKa = 10.63IMGYY21 pKa = 8.9VGEE24 pKa = 4.63MNFLSQWLIILEE36 pKa = 4.14TLILQLVLKK45 pKa = 6.77TQKK48 pKa = 10.91NNWTKK53 pKa = 10.67VPFKK57 pKa = 11.07QMLSKK62 pKa = 9.48FTQGIWRR69 pKa = 11.84NMSLISSFSSAKK81 pKa = 9.4CHH83 pKa = 6.24YY84 pKa = 10.29SLKK87 pKa = 10.44CWLKK91 pKa = 10.96YY92 pKa = 8.82ITT94 pKa = 4.65
Molecular weight: 11.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2552 |
94 |
643 |
364.6 |
40.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.564 ± 0.517 | 2.704 ± 0.718 |
6.348 ± 0.466 | 5.447 ± 0.279 |
4.35 ± 0.312 | 6.74 ± 0.411 |
2.743 ± 0.771 | 4.506 ± 0.4 |
5.486 ± 0.845 | 9.326 ± 0.57 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.685 ± 0.323 | 4.467 ± 0.585 |
6.583 ± 0.892 | 4.389 ± 0.447 |
4.859 ± 0.242 | 7.876 ± 0.892 |
6.74 ± 0.416 | 6.034 ± 0.386 |
1.528 ± 0.339 | 2.625 ± 0.387 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
