
Oricola thermophila
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Phyllobacteriaceae; Oricola
Average proteome isoelectric point is 6.24
Get precalculated fractions of proteins
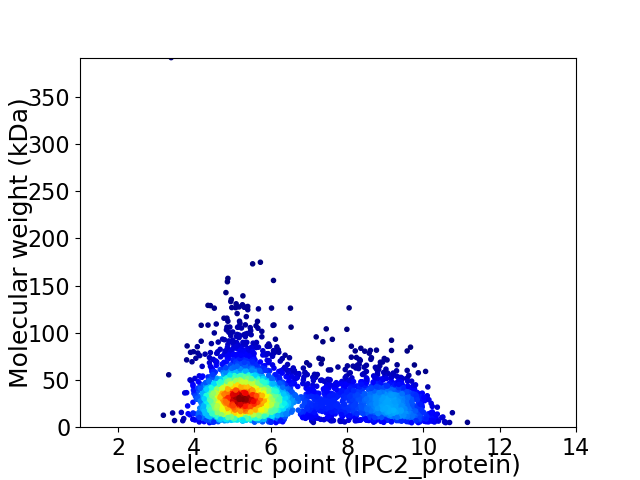
Virtual 2D-PAGE plot for 3798 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6N1VBZ1|A0A6N1VBZ1_9RHIZ TRAP transporter substrate-binding protein OS=Oricola thermophila OX=2742145 GN=HTY61_00350 PE=3 SV=1
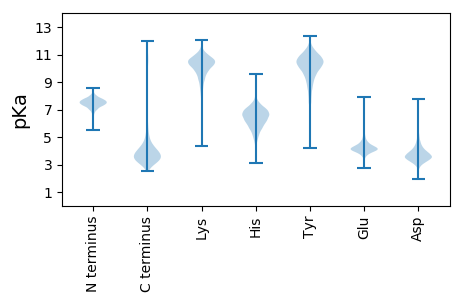
MM1 pKa = 7.64KK2 pKa = 10.21IKK4 pKa = 10.74SLLLGSAAALVAVSGARR21 pKa = 11.84AADD24 pKa = 3.45AVIIPEE30 pKa = 4.26PEE32 pKa = 3.59MVEE35 pKa = 4.04YY36 pKa = 11.19VRR38 pKa = 11.84VCDD41 pKa = 4.04AAGTGYY47 pKa = 10.49FYY49 pKa = 10.59IPGTEE54 pKa = 3.88TCLKK58 pKa = 9.35ISGFVRR64 pKa = 11.84YY65 pKa = 10.02QMDD68 pKa = 3.77FAGGDD73 pKa = 3.71GYY75 pKa = 11.49GDD77 pKa = 3.17QDD79 pKa = 3.54GWDD82 pKa = 3.44KK83 pKa = 11.3EE84 pKa = 4.22YY85 pKa = 11.33DD86 pKa = 3.62VNLLFEE92 pKa = 4.66AWNDD96 pKa = 3.58TEE98 pKa = 4.62YY99 pKa = 11.55GPLYY103 pKa = 10.18SQIRR107 pKa = 11.84LEE109 pKa = 4.15ADD111 pKa = 3.12GGVGEE116 pKa = 4.75DD117 pKa = 4.07ARR119 pKa = 11.84PLGDD123 pKa = 3.45SFVNIDD129 pKa = 3.13KK130 pKa = 10.96AYY132 pKa = 7.34FTLAGVTMGYY142 pKa = 10.54KK143 pKa = 10.32NSFWDD148 pKa = 3.55YY149 pKa = 11.28GFGGPADD156 pKa = 3.1IWNNGGPMTGMVGYY170 pKa = 7.21TANAGGFSISLALEE184 pKa = 4.61DD185 pKa = 5.51DD186 pKa = 5.03GSADD190 pKa = 3.39WTPAILGVVSGSIGDD205 pKa = 3.86LALTLGAVYY214 pKa = 10.22EE215 pKa = 4.52DD216 pKa = 4.86DD217 pKa = 4.07GATGEE222 pKa = 4.14VSDD225 pKa = 4.29WGFKK229 pKa = 8.63ATAAYY234 pKa = 10.79DD235 pKa = 3.34MGAAAVGFGVQYY247 pKa = 10.7SGEE250 pKa = 4.19TSGVNGSRR258 pKa = 11.84YY259 pKa = 5.24MTAYY263 pKa = 10.49DD264 pKa = 3.54WVLGADD270 pKa = 3.5VSFDD274 pKa = 3.36ATEE277 pKa = 4.04KK278 pKa = 10.96LSLGLGAQYY287 pKa = 10.93AINEE291 pKa = 4.3GGATDD296 pKa = 4.74DD297 pKa = 3.68DD298 pKa = 4.13WGIGAVASYY307 pKa = 10.73DD308 pKa = 3.38IVSGLNTEE316 pKa = 4.56LRR318 pKa = 11.84LRR320 pKa = 11.84WLEE323 pKa = 4.04DD324 pKa = 2.93TSTADD329 pKa = 4.65AVDD332 pKa = 3.56TSGWNARR339 pKa = 11.84LRR341 pKa = 11.84FTRR344 pKa = 11.84SFF346 pKa = 3.27
MM1 pKa = 7.64KK2 pKa = 10.21IKK4 pKa = 10.74SLLLGSAAALVAVSGARR21 pKa = 11.84AADD24 pKa = 3.45AVIIPEE30 pKa = 4.26PEE32 pKa = 3.59MVEE35 pKa = 4.04YY36 pKa = 11.19VRR38 pKa = 11.84VCDD41 pKa = 4.04AAGTGYY47 pKa = 10.49FYY49 pKa = 10.59IPGTEE54 pKa = 3.88TCLKK58 pKa = 9.35ISGFVRR64 pKa = 11.84YY65 pKa = 10.02QMDD68 pKa = 3.77FAGGDD73 pKa = 3.71GYY75 pKa = 11.49GDD77 pKa = 3.17QDD79 pKa = 3.54GWDD82 pKa = 3.44KK83 pKa = 11.3EE84 pKa = 4.22YY85 pKa = 11.33DD86 pKa = 3.62VNLLFEE92 pKa = 4.66AWNDD96 pKa = 3.58TEE98 pKa = 4.62YY99 pKa = 11.55GPLYY103 pKa = 10.18SQIRR107 pKa = 11.84LEE109 pKa = 4.15ADD111 pKa = 3.12GGVGEE116 pKa = 4.75DD117 pKa = 4.07ARR119 pKa = 11.84PLGDD123 pKa = 3.45SFVNIDD129 pKa = 3.13KK130 pKa = 10.96AYY132 pKa = 7.34FTLAGVTMGYY142 pKa = 10.54KK143 pKa = 10.32NSFWDD148 pKa = 3.55YY149 pKa = 11.28GFGGPADD156 pKa = 3.1IWNNGGPMTGMVGYY170 pKa = 7.21TANAGGFSISLALEE184 pKa = 4.61DD185 pKa = 5.51DD186 pKa = 5.03GSADD190 pKa = 3.39WTPAILGVVSGSIGDD205 pKa = 3.86LALTLGAVYY214 pKa = 10.22EE215 pKa = 4.52DD216 pKa = 4.86DD217 pKa = 4.07GATGEE222 pKa = 4.14VSDD225 pKa = 4.29WGFKK229 pKa = 8.63ATAAYY234 pKa = 10.79DD235 pKa = 3.34MGAAAVGFGVQYY247 pKa = 10.7SGEE250 pKa = 4.19TSGVNGSRR258 pKa = 11.84YY259 pKa = 5.24MTAYY263 pKa = 10.49DD264 pKa = 3.54WVLGADD270 pKa = 3.5VSFDD274 pKa = 3.36ATEE277 pKa = 4.04KK278 pKa = 10.96LSLGLGAQYY287 pKa = 10.93AINEE291 pKa = 4.3GGATDD296 pKa = 4.74DD297 pKa = 3.68DD298 pKa = 4.13WGIGAVASYY307 pKa = 10.73DD308 pKa = 3.38IVSGLNTEE316 pKa = 4.56LRR318 pKa = 11.84LRR320 pKa = 11.84WLEE323 pKa = 4.04DD324 pKa = 2.93TSTADD329 pKa = 4.65AVDD332 pKa = 3.56TSGWNARR339 pKa = 11.84LRR341 pKa = 11.84FTRR344 pKa = 11.84SFF346 pKa = 3.27
Molecular weight: 36.58 kDa
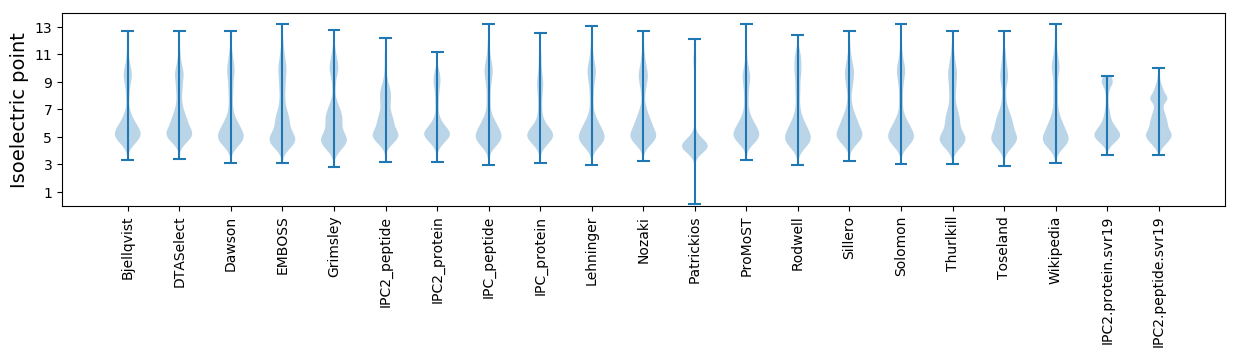
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6N1VJK5|A0A6N1VJK5_9RHIZ TetR/AcrR family transcriptional regulator OS=Oricola thermophila OX=2742145 GN=HTY61_14655 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.05VLAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.05VLAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1200695 |
41 |
3774 |
316.1 |
34.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.257 ± 0.052 | 0.845 ± 0.012 |
5.936 ± 0.036 | 6.383 ± 0.041 |
3.887 ± 0.026 | 8.69 ± 0.04 |
1.996 ± 0.018 | 5.506 ± 0.027 |
3.215 ± 0.033 | 9.539 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.669 ± 0.019 | 2.674 ± 0.021 |
4.979 ± 0.029 | 2.645 ± 0.022 |
7.255 ± 0.048 | 5.271 ± 0.032 |
5.179 ± 0.026 | 7.595 ± 0.035 |
1.267 ± 0.018 | 2.211 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
