
Faeces associated gemycircularvirus 12
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemytondvirus; Gemytondvirus ostri1; Ostrich associated gemytondvirus 1
Average proteome isoelectric point is 8.19
Get precalculated fractions of proteins
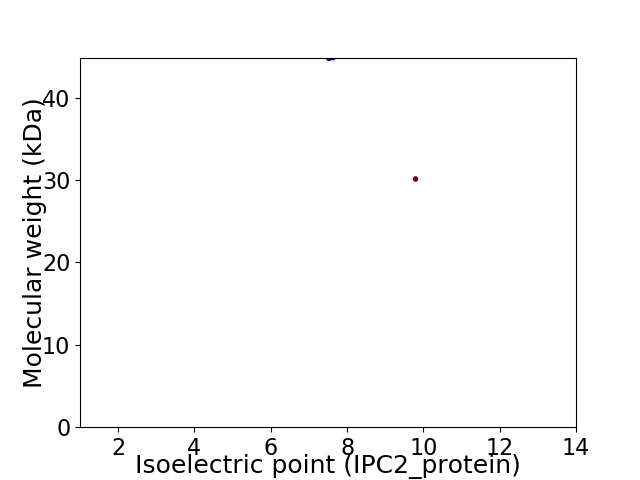
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|T1YRW7|T1YRW7_9VIRU Replication associated protein OS=Faeces associated gemycircularvirus 12 OX=1391027 PE=3 SV=1
MM1 pKa = 8.04PFRR4 pKa = 11.84FDD6 pKa = 3.25NKK8 pKa = 10.33RR9 pKa = 11.84VFLTYY14 pKa = 10.34SQVGQRR20 pKa = 11.84TLDD23 pKa = 3.54SIYY26 pKa = 11.23DD27 pKa = 3.65FLSTLKK33 pKa = 10.55DD34 pKa = 3.47GRR36 pKa = 11.84GEE38 pKa = 3.98LVAIEE43 pKa = 4.56YY44 pKa = 10.29IVVARR49 pKa = 11.84EE50 pKa = 3.54RR51 pKa = 11.84HH52 pKa = 5.47LDD54 pKa = 3.57GGFHH58 pKa = 5.88FHH60 pKa = 7.15AFILFRR66 pKa = 11.84EE67 pKa = 4.77RR68 pKa = 11.84FQSRR72 pKa = 11.84NQRR75 pKa = 11.84IFDD78 pKa = 3.75FDD80 pKa = 3.72GLHH83 pKa = 6.62PNIEE87 pKa = 4.43SVRR90 pKa = 11.84GEE92 pKa = 3.69RR93 pKa = 11.84NVANKK98 pKa = 8.38ITYY101 pKa = 7.06TKK103 pKa = 10.67KK104 pKa = 9.76DD105 pKa = 3.47GEE107 pKa = 4.42FKK109 pKa = 10.75EE110 pKa = 5.41HH111 pKa = 6.83GDD113 pKa = 3.66EE114 pKa = 4.01PTVAEE119 pKa = 4.28RR120 pKa = 11.84ANKK123 pKa = 7.67QQHH126 pKa = 5.59WLDD129 pKa = 5.09LIDD132 pKa = 4.53SSTDD136 pKa = 2.97AADD139 pKa = 4.01FMGKK143 pKa = 9.82AKK145 pKa = 10.34SVAPRR150 pKa = 11.84DD151 pKa = 3.62FVLQNDD157 pKa = 3.53KK158 pKa = 11.36FEE160 pKa = 4.44AFARR164 pKa = 11.84KK165 pKa = 9.49YY166 pKa = 8.37YY167 pKa = 10.98NNVATYY173 pKa = 9.11VSEE176 pKa = 4.15YY177 pKa = 10.38GPEE180 pKa = 4.13DD181 pKa = 3.28FVVPAEE187 pKa = 4.3CNDD190 pKa = 3.19WVRR193 pKa = 11.84DD194 pKa = 3.83VLNEE198 pKa = 3.8VCFLGTCYY206 pKa = 10.74SQIPNSNIVRR216 pKa = 11.84ILGTRR221 pKa = 11.84SSQMLGPHH229 pKa = 5.7RR230 pKa = 11.84RR231 pKa = 11.84RR232 pKa = 11.84RR233 pKa = 11.84TRR235 pKa = 11.84QTEE238 pKa = 3.81WARR241 pKa = 11.84CLGDD245 pKa = 3.65HH246 pKa = 7.43FYY248 pKa = 11.62NSTWFNLDD256 pKa = 3.07EE257 pKa = 4.4VDD259 pKa = 3.7PSKK262 pKa = 11.02KK263 pKa = 10.19YY264 pKa = 11.01AVFDD268 pKa = 5.09DD269 pKa = 3.12IRR271 pKa = 11.84LNEE274 pKa = 3.79KK275 pKa = 10.54SYY277 pKa = 10.51WSWKK281 pKa = 8.96PWFGGQKK288 pKa = 10.07EE289 pKa = 4.19FTVTDD294 pKa = 3.68KK295 pKa = 10.81FRR297 pKa = 11.84HH298 pKa = 5.28KK299 pKa = 10.47RR300 pKa = 11.84KK301 pKa = 10.21LMWGKK306 pKa = 9.69PIIWVCNPASDD317 pKa = 3.99PRR319 pKa = 11.84NIAGLDD325 pKa = 3.35SAKK328 pKa = 10.25FAEE331 pKa = 4.52SNPRR335 pKa = 11.84CARR338 pKa = 11.84RR339 pKa = 11.84AQEE342 pKa = 4.83IEE344 pKa = 5.16LILCNTPNPNGMFCEE359 pKa = 4.14DD360 pKa = 4.23VVCILYY366 pKa = 10.11IFVIHH371 pKa = 6.55RR372 pKa = 11.84VGPIKK377 pKa = 10.49PPSNYY382 pKa = 9.53IGG384 pKa = 3.42
MM1 pKa = 8.04PFRR4 pKa = 11.84FDD6 pKa = 3.25NKK8 pKa = 10.33RR9 pKa = 11.84VFLTYY14 pKa = 10.34SQVGQRR20 pKa = 11.84TLDD23 pKa = 3.54SIYY26 pKa = 11.23DD27 pKa = 3.65FLSTLKK33 pKa = 10.55DD34 pKa = 3.47GRR36 pKa = 11.84GEE38 pKa = 3.98LVAIEE43 pKa = 4.56YY44 pKa = 10.29IVVARR49 pKa = 11.84EE50 pKa = 3.54RR51 pKa = 11.84HH52 pKa = 5.47LDD54 pKa = 3.57GGFHH58 pKa = 5.88FHH60 pKa = 7.15AFILFRR66 pKa = 11.84EE67 pKa = 4.77RR68 pKa = 11.84FQSRR72 pKa = 11.84NQRR75 pKa = 11.84IFDD78 pKa = 3.75FDD80 pKa = 3.72GLHH83 pKa = 6.62PNIEE87 pKa = 4.43SVRR90 pKa = 11.84GEE92 pKa = 3.69RR93 pKa = 11.84NVANKK98 pKa = 8.38ITYY101 pKa = 7.06TKK103 pKa = 10.67KK104 pKa = 9.76DD105 pKa = 3.47GEE107 pKa = 4.42FKK109 pKa = 10.75EE110 pKa = 5.41HH111 pKa = 6.83GDD113 pKa = 3.66EE114 pKa = 4.01PTVAEE119 pKa = 4.28RR120 pKa = 11.84ANKK123 pKa = 7.67QQHH126 pKa = 5.59WLDD129 pKa = 5.09LIDD132 pKa = 4.53SSTDD136 pKa = 2.97AADD139 pKa = 4.01FMGKK143 pKa = 9.82AKK145 pKa = 10.34SVAPRR150 pKa = 11.84DD151 pKa = 3.62FVLQNDD157 pKa = 3.53KK158 pKa = 11.36FEE160 pKa = 4.44AFARR164 pKa = 11.84KK165 pKa = 9.49YY166 pKa = 8.37YY167 pKa = 10.98NNVATYY173 pKa = 9.11VSEE176 pKa = 4.15YY177 pKa = 10.38GPEE180 pKa = 4.13DD181 pKa = 3.28FVVPAEE187 pKa = 4.3CNDD190 pKa = 3.19WVRR193 pKa = 11.84DD194 pKa = 3.83VLNEE198 pKa = 3.8VCFLGTCYY206 pKa = 10.74SQIPNSNIVRR216 pKa = 11.84ILGTRR221 pKa = 11.84SSQMLGPHH229 pKa = 5.7RR230 pKa = 11.84RR231 pKa = 11.84RR232 pKa = 11.84RR233 pKa = 11.84TRR235 pKa = 11.84QTEE238 pKa = 3.81WARR241 pKa = 11.84CLGDD245 pKa = 3.65HH246 pKa = 7.43FYY248 pKa = 11.62NSTWFNLDD256 pKa = 3.07EE257 pKa = 4.4VDD259 pKa = 3.7PSKK262 pKa = 11.02KK263 pKa = 10.19YY264 pKa = 11.01AVFDD268 pKa = 5.09DD269 pKa = 3.12IRR271 pKa = 11.84LNEE274 pKa = 3.79KK275 pKa = 10.54SYY277 pKa = 10.51WSWKK281 pKa = 8.96PWFGGQKK288 pKa = 10.07EE289 pKa = 4.19FTVTDD294 pKa = 3.68KK295 pKa = 10.81FRR297 pKa = 11.84HH298 pKa = 5.28KK299 pKa = 10.47RR300 pKa = 11.84KK301 pKa = 10.21LMWGKK306 pKa = 9.69PIIWVCNPASDD317 pKa = 3.99PRR319 pKa = 11.84NIAGLDD325 pKa = 3.35SAKK328 pKa = 10.25FAEE331 pKa = 4.52SNPRR335 pKa = 11.84CARR338 pKa = 11.84RR339 pKa = 11.84AQEE342 pKa = 4.83IEE344 pKa = 5.16LILCNTPNPNGMFCEE359 pKa = 4.14DD360 pKa = 4.23VVCILYY366 pKa = 10.11IFVIHH371 pKa = 6.55RR372 pKa = 11.84VGPIKK377 pKa = 10.49PPSNYY382 pKa = 9.53IGG384 pKa = 3.42
Molecular weight: 44.74 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|T1YRW7|T1YRW7_9VIRU Replication associated protein OS=Faeces associated gemycircularvirus 12 OX=1391027 PE=3 SV=1
MM1 pKa = 6.72YY2 pKa = 10.3RR3 pKa = 11.84KK4 pKa = 9.79GGKK7 pKa = 8.33YY8 pKa = 9.63KK9 pKa = 10.14KK10 pKa = 9.77RR11 pKa = 11.84SYY13 pKa = 10.79KK14 pKa = 9.26RR15 pKa = 11.84RR16 pKa = 11.84KK17 pKa = 7.78YY18 pKa = 10.88NKK20 pKa = 8.72NKK22 pKa = 9.63VSKK25 pKa = 10.31KK26 pKa = 6.7VWKK29 pKa = 10.05ASKK32 pKa = 10.04KK33 pKa = 8.95AAKK36 pKa = 9.34AQIYY40 pKa = 10.07RR41 pKa = 11.84MSEE44 pKa = 3.81TKK46 pKa = 10.43VIHH49 pKa = 4.95STGYY53 pKa = 9.89SSYY56 pKa = 8.85ITAAEE61 pKa = 4.12SQFLCALHH69 pKa = 6.56PFYY72 pKa = 11.18FMTNHH77 pKa = 5.4SRR79 pKa = 11.84EE80 pKa = 4.08AGLVGNEE87 pKa = 3.05FDD89 pKa = 4.25LRR91 pKa = 11.84YY92 pKa = 10.06FEE94 pKa = 5.26LQGSLNFDD102 pKa = 3.9GFSTALMSDD111 pKa = 2.56VWLRR115 pKa = 11.84ISLIRR120 pKa = 11.84TPEE123 pKa = 3.79YY124 pKa = 9.62WPNDD128 pKa = 2.85AGTPVSSTIPTSTYY142 pKa = 11.06ALPLSAEE149 pKa = 3.86PWFSKK154 pKa = 10.58FNSNSVKK161 pKa = 10.27VLQTKK166 pKa = 7.63TIRR169 pKa = 11.84LPGAAYY175 pKa = 10.32VGNQTSTNRR184 pKa = 11.84IHH186 pKa = 7.25KK187 pKa = 9.51YY188 pKa = 10.43KK189 pKa = 10.55NFKK192 pKa = 10.11IKK194 pKa = 9.89FRR196 pKa = 11.84KK197 pKa = 9.63LKK199 pKa = 9.96GKK201 pKa = 8.21KK202 pKa = 8.56TIMSNYY208 pKa = 9.59DD209 pKa = 3.27EE210 pKa = 5.67NIVAGAGRR218 pKa = 11.84RR219 pKa = 11.84IRR221 pKa = 11.84QGQFYY226 pKa = 10.01IIIHH230 pKa = 5.32MASVNAVSGTTSTDD244 pKa = 2.55IVRR247 pKa = 11.84RR248 pKa = 11.84WLDD251 pKa = 3.03WTYY254 pKa = 11.82SMYY257 pKa = 11.2YY258 pKa = 10.34KK259 pKa = 10.69DD260 pKa = 3.2II261 pKa = 4.69
MM1 pKa = 6.72YY2 pKa = 10.3RR3 pKa = 11.84KK4 pKa = 9.79GGKK7 pKa = 8.33YY8 pKa = 9.63KK9 pKa = 10.14KK10 pKa = 9.77RR11 pKa = 11.84SYY13 pKa = 10.79KK14 pKa = 9.26RR15 pKa = 11.84RR16 pKa = 11.84KK17 pKa = 7.78YY18 pKa = 10.88NKK20 pKa = 8.72NKK22 pKa = 9.63VSKK25 pKa = 10.31KK26 pKa = 6.7VWKK29 pKa = 10.05ASKK32 pKa = 10.04KK33 pKa = 8.95AAKK36 pKa = 9.34AQIYY40 pKa = 10.07RR41 pKa = 11.84MSEE44 pKa = 3.81TKK46 pKa = 10.43VIHH49 pKa = 4.95STGYY53 pKa = 9.89SSYY56 pKa = 8.85ITAAEE61 pKa = 4.12SQFLCALHH69 pKa = 6.56PFYY72 pKa = 11.18FMTNHH77 pKa = 5.4SRR79 pKa = 11.84EE80 pKa = 4.08AGLVGNEE87 pKa = 3.05FDD89 pKa = 4.25LRR91 pKa = 11.84YY92 pKa = 10.06FEE94 pKa = 5.26LQGSLNFDD102 pKa = 3.9GFSTALMSDD111 pKa = 2.56VWLRR115 pKa = 11.84ISLIRR120 pKa = 11.84TPEE123 pKa = 3.79YY124 pKa = 9.62WPNDD128 pKa = 2.85AGTPVSSTIPTSTYY142 pKa = 11.06ALPLSAEE149 pKa = 3.86PWFSKK154 pKa = 10.58FNSNSVKK161 pKa = 10.27VLQTKK166 pKa = 7.63TIRR169 pKa = 11.84LPGAAYY175 pKa = 10.32VGNQTSTNRR184 pKa = 11.84IHH186 pKa = 7.25KK187 pKa = 9.51YY188 pKa = 10.43KK189 pKa = 10.55NFKK192 pKa = 10.11IKK194 pKa = 9.89FRR196 pKa = 11.84KK197 pKa = 9.63LKK199 pKa = 9.96GKK201 pKa = 8.21KK202 pKa = 8.56TIMSNYY208 pKa = 9.59DD209 pKa = 3.27EE210 pKa = 5.67NIVAGAGRR218 pKa = 11.84RR219 pKa = 11.84IRR221 pKa = 11.84QGQFYY226 pKa = 10.01IIIHH230 pKa = 5.32MASVNAVSGTTSTDD244 pKa = 2.55IVRR247 pKa = 11.84RR248 pKa = 11.84WLDD251 pKa = 3.03WTYY254 pKa = 11.82SMYY257 pKa = 11.2YY258 pKa = 10.34KK259 pKa = 10.69DD260 pKa = 3.2II261 pKa = 4.69
Molecular weight: 30.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
645 |
261 |
384 |
322.5 |
37.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.202 ± 0.404 | 1.55 ± 0.679 |
5.426 ± 1.373 | 4.806 ± 1.013 |
6.047 ± 0.843 | 5.736 ± 0.006 |
2.326 ± 0.238 | 6.047 ± 0.272 |
7.597 ± 1.598 | 5.891 ± 0.084 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.86 ± 0.478 | 5.736 ± 0.217 |
4.186 ± 0.652 | 2.946 ± 0.153 |
7.597 ± 0.63 | 7.287 ± 1.556 |
5.426 ± 1.078 | 6.047 ± 0.62 |
2.326 ± 0.016 | 4.961 ± 1.126 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
