
Great Island virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Orbivirus
Average proteome isoelectric point is 7.32
Get precalculated fractions of proteins
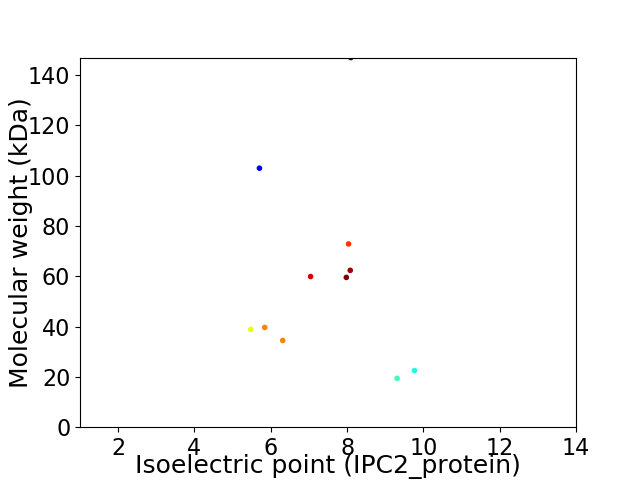
Virtual 2D-PAGE plot for 11 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E1AA97|E1AA97_9REOV VP6(Hel) OS=Great Island virus OX=204269 PE=4 SV=1
MM1 pKa = 7.38SADD4 pKa = 3.33KK5 pKa = 10.29QIPMIRR11 pKa = 11.84KK12 pKa = 7.78PFTRR16 pKa = 11.84TIVLKK21 pKa = 10.09STGKK25 pKa = 10.16DD26 pKa = 2.94DD27 pKa = 5.03YY28 pKa = 10.44VAKK31 pKa = 9.79MCNALGCEE39 pKa = 4.24YY40 pKa = 10.82LVVRR44 pKa = 11.84HH45 pKa = 5.59GLGMHH50 pKa = 6.11VAGSNTPTHH59 pKa = 6.29GCMLLLIPGAGSFRR73 pKa = 11.84LIDD76 pKa = 3.8RR77 pKa = 11.84DD78 pKa = 3.11QSTFVVVSSDD88 pKa = 3.21GLEE91 pKa = 4.14VQQDD95 pKa = 3.32RR96 pKa = 11.84WPGMRR101 pKa = 11.84FEE103 pKa = 4.71AVDD106 pKa = 3.42MFPRR110 pKa = 11.84CCRR113 pKa = 11.84IVVGDD118 pKa = 3.64VAADD122 pKa = 3.39SEE124 pKa = 4.51IRR126 pKa = 11.84FGSASGTVPPYY137 pKa = 9.66TAGEE141 pKa = 4.14SMEE144 pKa = 4.26AQDD147 pKa = 5.21EE148 pKa = 4.34ISLPGLDD155 pKa = 4.49FSVPDD160 pKa = 3.4SDD162 pKa = 3.94LRR164 pKa = 11.84EE165 pKa = 3.99YY166 pKa = 10.75RR167 pKa = 11.84EE168 pKa = 3.72KK169 pKa = 10.9LRR171 pKa = 11.84EE172 pKa = 4.09EE173 pKa = 4.19KK174 pKa = 10.53EE175 pKa = 3.76EE176 pKa = 3.7RR177 pKa = 11.84AAAILSALRR186 pKa = 11.84TTQQSRR192 pKa = 11.84TLGRR196 pKa = 11.84LHH198 pKa = 6.71GVKK201 pKa = 9.91TSDD204 pKa = 3.92LVTLRR209 pKa = 11.84PPQPPTPGRR218 pKa = 11.84NGHH221 pKa = 5.63SALMSSAPVPPPLVPTSSSTGSPALSTTGLKK252 pKa = 8.9RR253 pKa = 11.84TQKK256 pKa = 10.33LAPPPVAEE264 pKa = 4.29SAGPTFGADD273 pKa = 2.84SNRR276 pKa = 11.84FIEE279 pKa = 4.44EE280 pKa = 3.5ALEE283 pKa = 5.08AVMQDD288 pKa = 3.74DD289 pKa = 3.92ASGRR293 pKa = 11.84LNFGGEE299 pKa = 4.17PEE301 pKa = 4.38SVGLFSQNLGSFDD314 pKa = 3.95IPPSQLPAYY323 pKa = 9.6EE324 pKa = 3.93IDD326 pKa = 3.45EE327 pKa = 4.27TRR329 pKa = 11.84NKK331 pKa = 9.79YY332 pKa = 10.46KK333 pKa = 10.57YY334 pKa = 10.36VGMRR338 pKa = 11.84TSTRR342 pKa = 11.84LHH344 pKa = 6.8ALACDD349 pKa = 3.37DD350 pKa = 3.28RR351 pKa = 11.84VYY353 pKa = 10.66FVPSAEE359 pKa = 3.95
MM1 pKa = 7.38SADD4 pKa = 3.33KK5 pKa = 10.29QIPMIRR11 pKa = 11.84KK12 pKa = 7.78PFTRR16 pKa = 11.84TIVLKK21 pKa = 10.09STGKK25 pKa = 10.16DD26 pKa = 2.94DD27 pKa = 5.03YY28 pKa = 10.44VAKK31 pKa = 9.79MCNALGCEE39 pKa = 4.24YY40 pKa = 10.82LVVRR44 pKa = 11.84HH45 pKa = 5.59GLGMHH50 pKa = 6.11VAGSNTPTHH59 pKa = 6.29GCMLLLIPGAGSFRR73 pKa = 11.84LIDD76 pKa = 3.8RR77 pKa = 11.84DD78 pKa = 3.11QSTFVVVSSDD88 pKa = 3.21GLEE91 pKa = 4.14VQQDD95 pKa = 3.32RR96 pKa = 11.84WPGMRR101 pKa = 11.84FEE103 pKa = 4.71AVDD106 pKa = 3.42MFPRR110 pKa = 11.84CCRR113 pKa = 11.84IVVGDD118 pKa = 3.64VAADD122 pKa = 3.39SEE124 pKa = 4.51IRR126 pKa = 11.84FGSASGTVPPYY137 pKa = 9.66TAGEE141 pKa = 4.14SMEE144 pKa = 4.26AQDD147 pKa = 5.21EE148 pKa = 4.34ISLPGLDD155 pKa = 4.49FSVPDD160 pKa = 3.4SDD162 pKa = 3.94LRR164 pKa = 11.84EE165 pKa = 3.99YY166 pKa = 10.75RR167 pKa = 11.84EE168 pKa = 3.72KK169 pKa = 10.9LRR171 pKa = 11.84EE172 pKa = 4.09EE173 pKa = 4.19KK174 pKa = 10.53EE175 pKa = 3.76EE176 pKa = 3.7RR177 pKa = 11.84AAAILSALRR186 pKa = 11.84TTQQSRR192 pKa = 11.84TLGRR196 pKa = 11.84LHH198 pKa = 6.71GVKK201 pKa = 9.91TSDD204 pKa = 3.92LVTLRR209 pKa = 11.84PPQPPTPGRR218 pKa = 11.84NGHH221 pKa = 5.63SALMSSAPVPPPLVPTSSSTGSPALSTTGLKK252 pKa = 8.9RR253 pKa = 11.84TQKK256 pKa = 10.33LAPPPVAEE264 pKa = 4.29SAGPTFGADD273 pKa = 2.84SNRR276 pKa = 11.84FIEE279 pKa = 4.44EE280 pKa = 3.5ALEE283 pKa = 5.08AVMQDD288 pKa = 3.74DD289 pKa = 3.92ASGRR293 pKa = 11.84LNFGGEE299 pKa = 4.17PEE301 pKa = 4.38SVGLFSQNLGSFDD314 pKa = 3.95IPPSQLPAYY323 pKa = 9.6EE324 pKa = 3.93IDD326 pKa = 3.45EE327 pKa = 4.27TRR329 pKa = 11.84NKK331 pKa = 9.79YY332 pKa = 10.46KK333 pKa = 10.57YY334 pKa = 10.36VGMRR338 pKa = 11.84TSTRR342 pKa = 11.84LHH344 pKa = 6.8ALACDD349 pKa = 3.37DD350 pKa = 3.28RR351 pKa = 11.84VYY353 pKa = 10.66FVPSAEE359 pKa = 3.95
Molecular weight: 38.88 kDa
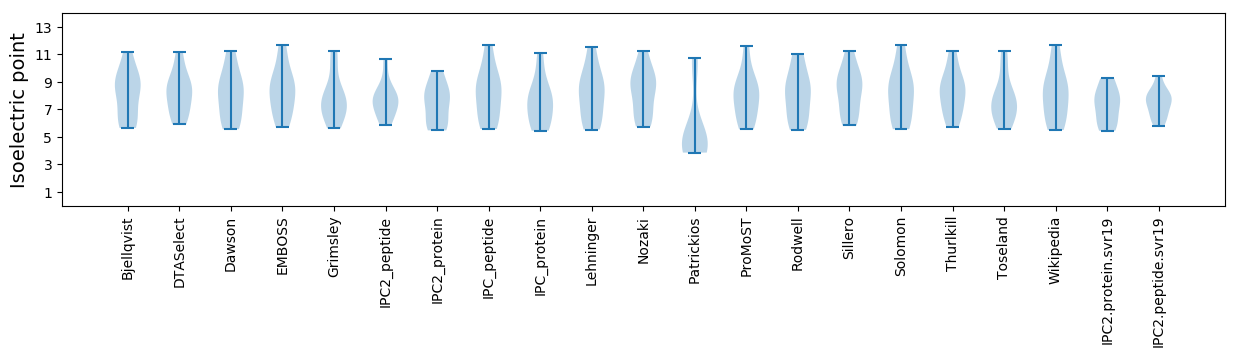
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E1AA99|E1AA99_9REOV NS3 OS=Great Island virus OX=204269 PE=4 SV=1
MM1 pKa = 7.39KK2 pKa = 10.32NEE4 pKa = 3.55KK5 pKa = 9.68AAYY8 pKa = 8.47GAASEE13 pKa = 4.25VLKK16 pKa = 10.88DD17 pKa = 4.17DD18 pKa = 3.45EE19 pKa = 4.57TTRR22 pKa = 11.84MLKK25 pKa = 8.71MQVNEE30 pKa = 4.22CSLTEE35 pKa = 3.85MRR37 pKa = 11.84QTYY40 pKa = 9.73QSLKK44 pKa = 9.35RR45 pKa = 11.84RR46 pKa = 11.84CRR48 pKa = 11.84LLYY51 pKa = 10.5YY52 pKa = 10.96GEE54 pKa = 4.6LLCLAAALGLTLVLMVPSASRR75 pKa = 11.84VLEE78 pKa = 4.31GALTQANITGHH89 pKa = 6.33VITGILTSLAIFLQHH104 pKa = 5.99HH105 pKa = 6.25RR106 pKa = 11.84ARR108 pKa = 11.84LLKK111 pKa = 10.05RR112 pKa = 11.84KK113 pKa = 9.72RR114 pKa = 11.84SIKK117 pKa = 9.65RR118 pKa = 11.84DD119 pKa = 2.86IVKK122 pKa = 10.4RR123 pKa = 11.84MTYY126 pKa = 9.85ISLARR131 pKa = 11.84RR132 pKa = 11.84MGSQFPEE139 pKa = 4.2SAGAGSDD146 pKa = 3.38FRR148 pKa = 11.84ARR150 pKa = 11.84LMALAEE156 pKa = 4.04EE157 pKa = 4.72AEE159 pKa = 4.29RR160 pKa = 11.84ARR162 pKa = 11.84DD163 pKa = 3.65DD164 pKa = 3.96SDD166 pKa = 2.75WRR168 pKa = 11.84RR169 pKa = 11.84WPP171 pKa = 3.41
MM1 pKa = 7.39KK2 pKa = 10.32NEE4 pKa = 3.55KK5 pKa = 9.68AAYY8 pKa = 8.47GAASEE13 pKa = 4.25VLKK16 pKa = 10.88DD17 pKa = 4.17DD18 pKa = 3.45EE19 pKa = 4.57TTRR22 pKa = 11.84MLKK25 pKa = 8.71MQVNEE30 pKa = 4.22CSLTEE35 pKa = 3.85MRR37 pKa = 11.84QTYY40 pKa = 9.73QSLKK44 pKa = 9.35RR45 pKa = 11.84RR46 pKa = 11.84CRR48 pKa = 11.84LLYY51 pKa = 10.5YY52 pKa = 10.96GEE54 pKa = 4.6LLCLAAALGLTLVLMVPSASRR75 pKa = 11.84VLEE78 pKa = 4.31GALTQANITGHH89 pKa = 6.33VITGILTSLAIFLQHH104 pKa = 5.99HH105 pKa = 6.25RR106 pKa = 11.84ARR108 pKa = 11.84LLKK111 pKa = 10.05RR112 pKa = 11.84KK113 pKa = 9.72RR114 pKa = 11.84SIKK117 pKa = 9.65RR118 pKa = 11.84DD119 pKa = 2.86IVKK122 pKa = 10.4RR123 pKa = 11.84MTYY126 pKa = 9.85ISLARR131 pKa = 11.84RR132 pKa = 11.84MGSQFPEE139 pKa = 4.2SAGAGSDD146 pKa = 3.38FRR148 pKa = 11.84ARR150 pKa = 11.84LMALAEE156 pKa = 4.04EE157 pKa = 4.72AEE159 pKa = 4.29RR160 pKa = 11.84ARR162 pKa = 11.84DD163 pKa = 3.65DD164 pKa = 3.96SDD166 pKa = 2.75WRR168 pKa = 11.84RR169 pKa = 11.84WPP171 pKa = 3.41
Molecular weight: 19.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5845 |
171 |
1285 |
531.4 |
59.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.393 ± 0.615 | 1.317 ± 0.203 |
5.8 ± 0.34 | 6.176 ± 0.344 |
3.49 ± 0.311 | 5.885 ± 0.387 |
2.789 ± 0.259 | 4.808 ± 0.302 |
2.515 ± 0.455 | 9.478 ± 0.349 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.652 ± 0.187 | 2.618 ± 0.266 |
5.287 ± 0.354 | 3.849 ± 0.329 |
10.197 ± 0.383 | 6.347 ± 0.377 |
5.954 ± 0.267 | 7.169 ± 0.224 |
1.112 ± 0.231 | 3.165 ± 0.239 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
