
Streptomyces sp. JV178
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces; unclassified Streptomyces
Average proteome isoelectric point is 6.32
Get precalculated fractions of proteins
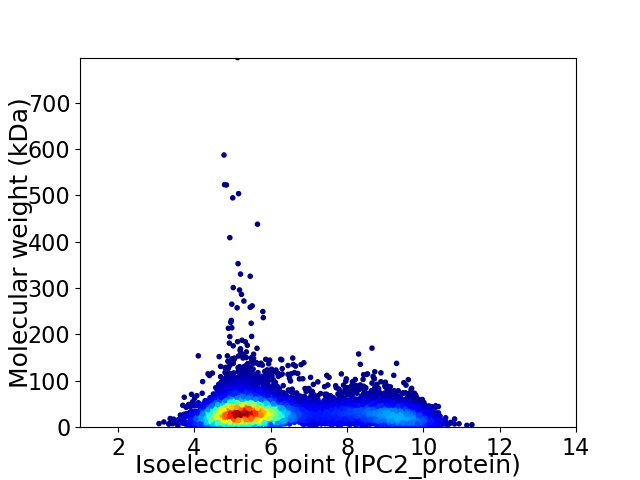
Virtual 2D-PAGE plot for 8845 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2G9DZ85|A0A2G9DZ85_9ACTN Iron permease OS=Streptomyces sp. JV178 OX=858632 GN=CTU88_05865 PE=3 SV=1
MM1 pKa = 7.81TDD3 pKa = 3.42DD4 pKa = 3.32QHH6 pKa = 9.59GEE8 pKa = 4.04GTPQQGGQSGWEE20 pKa = 3.83RR21 pKa = 11.84LPQGDD26 pKa = 4.38YY27 pKa = 11.43DD28 pKa = 4.65DD29 pKa = 4.95GATTFVQLPEE39 pKa = 4.39GGIDD43 pKa = 3.5ALFAADD49 pKa = 4.81SPLAAPGHH57 pKa = 6.43GYY59 pKa = 9.88VPPPIAPNPPGAAGTDD75 pKa = 3.67PSGTGSWAMPAEE87 pKa = 4.77GAQWHH92 pKa = 7.19DD93 pKa = 4.05PNAGQQQPPAQDD105 pKa = 2.8GFTYY109 pKa = 10.76NPGSTGQWTFEE120 pKa = 3.99EE121 pKa = 5.0PGAQEE126 pKa = 4.04GTAPGHH132 pKa = 6.56DD133 pKa = 3.71VTGEE137 pKa = 3.58WSIPVAGGDD146 pKa = 3.84LPDD149 pKa = 3.75EE150 pKa = 4.63SGEE153 pKa = 4.14FSTSALVEE161 pKa = 4.16QWGGTPPATLPGGAPAPWATEE182 pKa = 4.01QIGTAWTAPPPPTRR196 pKa = 11.84AEE198 pKa = 3.72EE199 pKa = 4.26SAAGEE204 pKa = 4.16PPADD208 pKa = 3.9ARR210 pKa = 11.84AEE212 pKa = 4.2APGEE216 pKa = 3.93PSSAPVEE223 pKa = 4.39APAPDD228 pKa = 3.62ASEE231 pKa = 4.22EE232 pKa = 4.26TPEE235 pKa = 4.22APEE238 pKa = 4.42GAQEE242 pKa = 4.16SAGGDD247 pKa = 3.55EE248 pKa = 4.45QAAEE252 pKa = 4.26ADD254 pKa = 3.97GDD256 pKa = 4.14STGPQEE262 pKa = 4.58PGTPAEE268 pKa = 4.52ASDD271 pKa = 4.89AEE273 pKa = 4.5TPDD276 pKa = 4.63AEE278 pKa = 4.71DD279 pKa = 4.6AGAADD284 pKa = 4.52PAAEE288 pKa = 4.24PTPPPHH294 pKa = 7.68DD295 pKa = 3.93DD296 pKa = 4.16HH297 pKa = 7.13PLASYY302 pKa = 10.27VLSVNGADD310 pKa = 5.33RR311 pKa = 11.84PVTDD315 pKa = 3.02AWIGEE320 pKa = 4.15SLLYY324 pKa = 10.18VLRR327 pKa = 11.84EE328 pKa = 3.94RR329 pKa = 11.84LGLAGAKK336 pKa = 9.72DD337 pKa = 3.75GCSQGEE343 pKa = 4.12CGACNVQVDD352 pKa = 3.94GRR354 pKa = 11.84LVASCLVPAVTAASSEE370 pKa = 3.91VRR372 pKa = 11.84TVEE375 pKa = 3.85GLAVDD380 pKa = 4.62GQPSDD385 pKa = 3.61VQRR388 pKa = 11.84ALARR392 pKa = 11.84CGAVQCGFCVPGMAMTVHH410 pKa = 7.27DD411 pKa = 5.16LLEE414 pKa = 4.78GNPAPTEE421 pKa = 4.03LEE423 pKa = 3.87ARR425 pKa = 11.84QALCGNLCRR434 pKa = 11.84CSGYY438 pKa = 10.31RR439 pKa = 11.84GVLAAVRR446 pKa = 11.84DD447 pKa = 4.42VVAEE451 pKa = 4.45RR452 pKa = 11.84AAHH455 pKa = 6.06AAGDD459 pKa = 4.1GDD461 pKa = 4.0TGADD465 pKa = 3.26TDD467 pKa = 3.87EE468 pKa = 4.58ARR470 pKa = 11.84IPHH473 pKa = 5.5QAGPGAGGVNPAAYY487 pKa = 7.67EE488 pKa = 4.12AQGTPPQVPNDD499 pKa = 3.84PGQSYY504 pKa = 10.58GGQGLSFGGQDD515 pKa = 3.54DD516 pKa = 4.89TYY518 pKa = 11.01GGQGQSFGGHH528 pKa = 6.22GDD530 pKa = 3.47TYY532 pKa = 11.11GGQGQSFGGQDD543 pKa = 3.23QSFGGQGDD551 pKa = 4.19TYY553 pKa = 10.89GGQGQSFGGHH563 pKa = 6.72DD564 pKa = 4.24DD565 pKa = 3.75SYY567 pKa = 11.69GGQGQSFGGHH577 pKa = 6.22GDD579 pKa = 3.47TYY581 pKa = 11.11GGQGQSFGGQDD592 pKa = 3.27QSFGGQDD599 pKa = 3.21QSFGGQDD606 pKa = 3.29DD607 pKa = 4.86TYY609 pKa = 11.01GGQGQSFGGHH619 pKa = 7.18DD620 pKa = 3.96DD621 pKa = 3.78TYY623 pKa = 11.43GGQGRR628 pKa = 11.84NFGDD632 pKa = 3.32QDD634 pKa = 3.36QSFGQDD640 pKa = 2.59GGQAA644 pKa = 3.31
MM1 pKa = 7.81TDD3 pKa = 3.42DD4 pKa = 3.32QHH6 pKa = 9.59GEE8 pKa = 4.04GTPQQGGQSGWEE20 pKa = 3.83RR21 pKa = 11.84LPQGDD26 pKa = 4.38YY27 pKa = 11.43DD28 pKa = 4.65DD29 pKa = 4.95GATTFVQLPEE39 pKa = 4.39GGIDD43 pKa = 3.5ALFAADD49 pKa = 4.81SPLAAPGHH57 pKa = 6.43GYY59 pKa = 9.88VPPPIAPNPPGAAGTDD75 pKa = 3.67PSGTGSWAMPAEE87 pKa = 4.77GAQWHH92 pKa = 7.19DD93 pKa = 4.05PNAGQQQPPAQDD105 pKa = 2.8GFTYY109 pKa = 10.76NPGSTGQWTFEE120 pKa = 3.99EE121 pKa = 5.0PGAQEE126 pKa = 4.04GTAPGHH132 pKa = 6.56DD133 pKa = 3.71VTGEE137 pKa = 3.58WSIPVAGGDD146 pKa = 3.84LPDD149 pKa = 3.75EE150 pKa = 4.63SGEE153 pKa = 4.14FSTSALVEE161 pKa = 4.16QWGGTPPATLPGGAPAPWATEE182 pKa = 4.01QIGTAWTAPPPPTRR196 pKa = 11.84AEE198 pKa = 3.72EE199 pKa = 4.26SAAGEE204 pKa = 4.16PPADD208 pKa = 3.9ARR210 pKa = 11.84AEE212 pKa = 4.2APGEE216 pKa = 3.93PSSAPVEE223 pKa = 4.39APAPDD228 pKa = 3.62ASEE231 pKa = 4.22EE232 pKa = 4.26TPEE235 pKa = 4.22APEE238 pKa = 4.42GAQEE242 pKa = 4.16SAGGDD247 pKa = 3.55EE248 pKa = 4.45QAAEE252 pKa = 4.26ADD254 pKa = 3.97GDD256 pKa = 4.14STGPQEE262 pKa = 4.58PGTPAEE268 pKa = 4.52ASDD271 pKa = 4.89AEE273 pKa = 4.5TPDD276 pKa = 4.63AEE278 pKa = 4.71DD279 pKa = 4.6AGAADD284 pKa = 4.52PAAEE288 pKa = 4.24PTPPPHH294 pKa = 7.68DD295 pKa = 3.93DD296 pKa = 4.16HH297 pKa = 7.13PLASYY302 pKa = 10.27VLSVNGADD310 pKa = 5.33RR311 pKa = 11.84PVTDD315 pKa = 3.02AWIGEE320 pKa = 4.15SLLYY324 pKa = 10.18VLRR327 pKa = 11.84EE328 pKa = 3.94RR329 pKa = 11.84LGLAGAKK336 pKa = 9.72DD337 pKa = 3.75GCSQGEE343 pKa = 4.12CGACNVQVDD352 pKa = 3.94GRR354 pKa = 11.84LVASCLVPAVTAASSEE370 pKa = 3.91VRR372 pKa = 11.84TVEE375 pKa = 3.85GLAVDD380 pKa = 4.62GQPSDD385 pKa = 3.61VQRR388 pKa = 11.84ALARR392 pKa = 11.84CGAVQCGFCVPGMAMTVHH410 pKa = 7.27DD411 pKa = 5.16LLEE414 pKa = 4.78GNPAPTEE421 pKa = 4.03LEE423 pKa = 3.87ARR425 pKa = 11.84QALCGNLCRR434 pKa = 11.84CSGYY438 pKa = 10.31RR439 pKa = 11.84GVLAAVRR446 pKa = 11.84DD447 pKa = 4.42VVAEE451 pKa = 4.45RR452 pKa = 11.84AAHH455 pKa = 6.06AAGDD459 pKa = 4.1GDD461 pKa = 4.0TGADD465 pKa = 3.26TDD467 pKa = 3.87EE468 pKa = 4.58ARR470 pKa = 11.84IPHH473 pKa = 5.5QAGPGAGGVNPAAYY487 pKa = 7.67EE488 pKa = 4.12AQGTPPQVPNDD499 pKa = 3.84PGQSYY504 pKa = 10.58GGQGLSFGGQDD515 pKa = 3.54DD516 pKa = 4.89TYY518 pKa = 11.01GGQGQSFGGHH528 pKa = 6.22GDD530 pKa = 3.47TYY532 pKa = 11.11GGQGQSFGGQDD543 pKa = 3.23QSFGGQGDD551 pKa = 4.19TYY553 pKa = 10.89GGQGQSFGGHH563 pKa = 6.72DD564 pKa = 4.24DD565 pKa = 3.75SYY567 pKa = 11.69GGQGQSFGGHH577 pKa = 6.22GDD579 pKa = 3.47TYY581 pKa = 11.11GGQGQSFGGQDD592 pKa = 3.27QSFGGQDD599 pKa = 3.21QSFGGQDD606 pKa = 3.29DD607 pKa = 4.86TYY609 pKa = 11.01GGQGQSFGGHH619 pKa = 7.18DD620 pKa = 3.96DD621 pKa = 3.78TYY623 pKa = 11.43GGQGRR628 pKa = 11.84NFGDD632 pKa = 3.32QDD634 pKa = 3.36QSFGQDD640 pKa = 2.59GGQAA644 pKa = 3.31
Molecular weight: 64.6 kDa
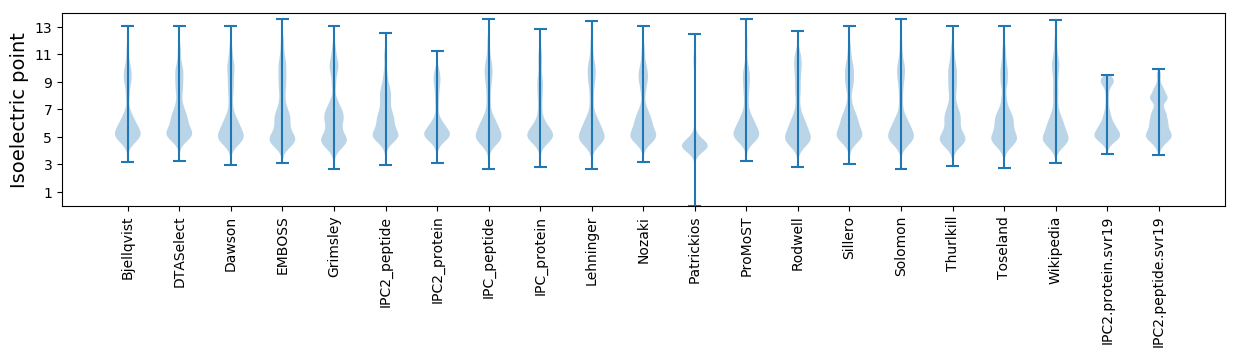
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2G9DQE3|A0A2G9DQE3_9ACTN Uncharacterized protein OS=Streptomyces sp. JV178 OX=858632 GN=CTU88_19325 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILANRR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.46GRR40 pKa = 11.84ASLSAA45 pKa = 3.83
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILANRR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.46GRR40 pKa = 11.84ASLSAA45 pKa = 3.83
Molecular weight: 5.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2953601 |
26 |
7399 |
333.9 |
35.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.31 ± 0.037 | 0.779 ± 0.008 |
6.164 ± 0.02 | 5.689 ± 0.028 |
2.729 ± 0.014 | 9.524 ± 0.03 |
2.328 ± 0.014 | 3.1 ± 0.019 |
2.199 ± 0.024 | 10.231 ± 0.034 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.689 ± 0.01 | 1.827 ± 0.015 |
6.144 ± 0.026 | 2.669 ± 0.016 |
8.031 ± 0.031 | 5.078 ± 0.021 |
6.358 ± 0.026 | 8.439 ± 0.025 |
1.573 ± 0.011 | 2.138 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
