
Salmonella phage IKe (Bacteriophage IKe)
Taxonomy: Viruses; Monodnaviria; Loebvirae; Hofneiviricota; Faserviricetes; Tubulavirales; Inoviridae; Lineavirus; Salmonella virus IKe
Average proteome isoelectric point is 7.07
Get precalculated fractions of proteins
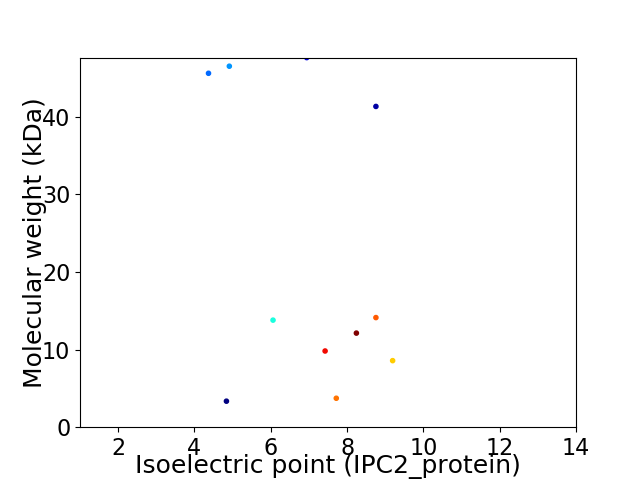
Virtual 2D-PAGE plot for 11 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P03667|G4P_BPIKE Virion export protein OS=Salmonella phage IKe OX=10867 GN=IV PE=3 SV=1
MM1 pKa = 7.15KK2 pKa = 10.05RR3 pKa = 11.84KK4 pKa = 9.92IIAISLFLYY13 pKa = 10.26IPLSNADD20 pKa = 3.16NWEE23 pKa = 4.73SITKK27 pKa = 10.09SYY29 pKa = 9.41YY30 pKa = 9.4TGFAISKK37 pKa = 8.02TVEE40 pKa = 4.13SKK42 pKa = 11.22DD43 pKa = 3.5KK44 pKa = 10.88DD45 pKa = 3.6GKK47 pKa = 9.15PVRR50 pKa = 11.84KK51 pKa = 9.06EE52 pKa = 4.03VITQADD58 pKa = 4.19LTTACNDD65 pKa = 3.48AKK67 pKa = 11.1ASAQNVFNQIKK78 pKa = 8.71LTLSGTWPNSQFRR91 pKa = 11.84LVTGDD96 pKa = 3.27TCVYY100 pKa = 10.11NGSPGEE106 pKa = 4.16KK107 pKa = 8.83TEE109 pKa = 3.93SWSIRR114 pKa = 11.84AQVEE118 pKa = 3.63GDD120 pKa = 3.29IQRR123 pKa = 11.84SVPDD127 pKa = 4.24EE128 pKa = 4.42EE129 pKa = 5.3PSEE132 pKa = 4.06QTPEE136 pKa = 4.36EE137 pKa = 3.99ICEE140 pKa = 4.15AKK142 pKa = 10.56PPIDD146 pKa = 3.35GVFNNVFKK154 pKa = 11.02GDD156 pKa = 3.35EE157 pKa = 3.98GGFYY161 pKa = 10.31INYY164 pKa = 9.6NGCEE168 pKa = 3.98YY169 pKa = 10.28EE170 pKa = 4.12ATGVTVCQNDD180 pKa = 3.77GTVCSSSAWKK190 pKa = 7.96PTGYY194 pKa = 10.11VPEE197 pKa = 4.74SGEE200 pKa = 4.13PSSSPLKK207 pKa = 11.08DD208 pKa = 3.02GDD210 pKa = 3.73TGGTGEE216 pKa = 4.86GGSDD220 pKa = 2.97TGGDD224 pKa = 3.31TGGGDD229 pKa = 3.38TGGGSTGGDD238 pKa = 3.1TGGSSGGGSSGGGSSGGSTGKK259 pKa = 10.44SLTKK263 pKa = 10.41EE264 pKa = 3.95DD265 pKa = 3.36VTAAIHH271 pKa = 5.43VASPSIGDD279 pKa = 3.69AVKK282 pKa = 10.86DD283 pKa = 4.01SLTEE287 pKa = 5.58DD288 pKa = 3.1NDD290 pKa = 3.84QYY292 pKa = 12.07DD293 pKa = 4.0NQKK296 pKa = 10.74KK297 pKa = 10.41ADD299 pKa = 3.92EE300 pKa = 4.18QSAKK304 pKa = 10.72ASASVSDD311 pKa = 5.04AISDD315 pKa = 3.64GMRR318 pKa = 11.84GVGNFVDD325 pKa = 4.54DD326 pKa = 4.6FGGEE330 pKa = 3.92SSQYY334 pKa = 8.8GTGNSEE340 pKa = 3.81MDD342 pKa = 3.99LSVSLAKK349 pKa = 10.52GQLGIDD355 pKa = 3.73RR356 pKa = 11.84EE357 pKa = 4.7GHH359 pKa = 4.86GSAWEE364 pKa = 4.1SFLNDD369 pKa = 3.12GALRR373 pKa = 11.84PSIPTGHH380 pKa = 6.79GCTNFVMYY388 pKa = 10.14QGSVYY393 pKa = 10.25QIEE396 pKa = 4.31IGCDD400 pKa = 2.93KK401 pKa = 11.26LNDD404 pKa = 3.72IKK406 pKa = 11.3SVLSWVMYY414 pKa = 10.54CLTFWYY420 pKa = 10.02VFQSVTSLLRR430 pKa = 11.84KK431 pKa = 10.08GEE433 pKa = 3.98QQ434 pKa = 2.97
MM1 pKa = 7.15KK2 pKa = 10.05RR3 pKa = 11.84KK4 pKa = 9.92IIAISLFLYY13 pKa = 10.26IPLSNADD20 pKa = 3.16NWEE23 pKa = 4.73SITKK27 pKa = 10.09SYY29 pKa = 9.41YY30 pKa = 9.4TGFAISKK37 pKa = 8.02TVEE40 pKa = 4.13SKK42 pKa = 11.22DD43 pKa = 3.5KK44 pKa = 10.88DD45 pKa = 3.6GKK47 pKa = 9.15PVRR50 pKa = 11.84KK51 pKa = 9.06EE52 pKa = 4.03VITQADD58 pKa = 4.19LTTACNDD65 pKa = 3.48AKK67 pKa = 11.1ASAQNVFNQIKK78 pKa = 8.71LTLSGTWPNSQFRR91 pKa = 11.84LVTGDD96 pKa = 3.27TCVYY100 pKa = 10.11NGSPGEE106 pKa = 4.16KK107 pKa = 8.83TEE109 pKa = 3.93SWSIRR114 pKa = 11.84AQVEE118 pKa = 3.63GDD120 pKa = 3.29IQRR123 pKa = 11.84SVPDD127 pKa = 4.24EE128 pKa = 4.42EE129 pKa = 5.3PSEE132 pKa = 4.06QTPEE136 pKa = 4.36EE137 pKa = 3.99ICEE140 pKa = 4.15AKK142 pKa = 10.56PPIDD146 pKa = 3.35GVFNNVFKK154 pKa = 11.02GDD156 pKa = 3.35EE157 pKa = 3.98GGFYY161 pKa = 10.31INYY164 pKa = 9.6NGCEE168 pKa = 3.98YY169 pKa = 10.28EE170 pKa = 4.12ATGVTVCQNDD180 pKa = 3.77GTVCSSSAWKK190 pKa = 7.96PTGYY194 pKa = 10.11VPEE197 pKa = 4.74SGEE200 pKa = 4.13PSSSPLKK207 pKa = 11.08DD208 pKa = 3.02GDD210 pKa = 3.73TGGTGEE216 pKa = 4.86GGSDD220 pKa = 2.97TGGDD224 pKa = 3.31TGGGDD229 pKa = 3.38TGGGSTGGDD238 pKa = 3.1TGGSSGGGSSGGGSSGGSTGKK259 pKa = 10.44SLTKK263 pKa = 10.41EE264 pKa = 3.95DD265 pKa = 3.36VTAAIHH271 pKa = 5.43VASPSIGDD279 pKa = 3.69AVKK282 pKa = 10.86DD283 pKa = 4.01SLTEE287 pKa = 5.58DD288 pKa = 3.1NDD290 pKa = 3.84QYY292 pKa = 12.07DD293 pKa = 4.0NQKK296 pKa = 10.74KK297 pKa = 10.41ADD299 pKa = 3.92EE300 pKa = 4.18QSAKK304 pKa = 10.72ASASVSDD311 pKa = 5.04AISDD315 pKa = 3.64GMRR318 pKa = 11.84GVGNFVDD325 pKa = 4.54DD326 pKa = 4.6FGGEE330 pKa = 3.92SSQYY334 pKa = 8.8GTGNSEE340 pKa = 3.81MDD342 pKa = 3.99LSVSLAKK349 pKa = 10.52GQLGIDD355 pKa = 3.73RR356 pKa = 11.84EE357 pKa = 4.7GHH359 pKa = 4.86GSAWEE364 pKa = 4.1SFLNDD369 pKa = 3.12GALRR373 pKa = 11.84PSIPTGHH380 pKa = 6.79GCTNFVMYY388 pKa = 10.14QGSVYY393 pKa = 10.25QIEE396 pKa = 4.31IGCDD400 pKa = 2.93KK401 pKa = 11.26LNDD404 pKa = 3.72IKK406 pKa = 11.3SVLSWVMYY414 pKa = 10.54CLTFWYY420 pKa = 10.02VFQSVTSLLRR430 pKa = 11.84KK431 pKa = 10.08GEE433 pKa = 3.98QQ434 pKa = 2.97
Molecular weight: 45.57 kDa
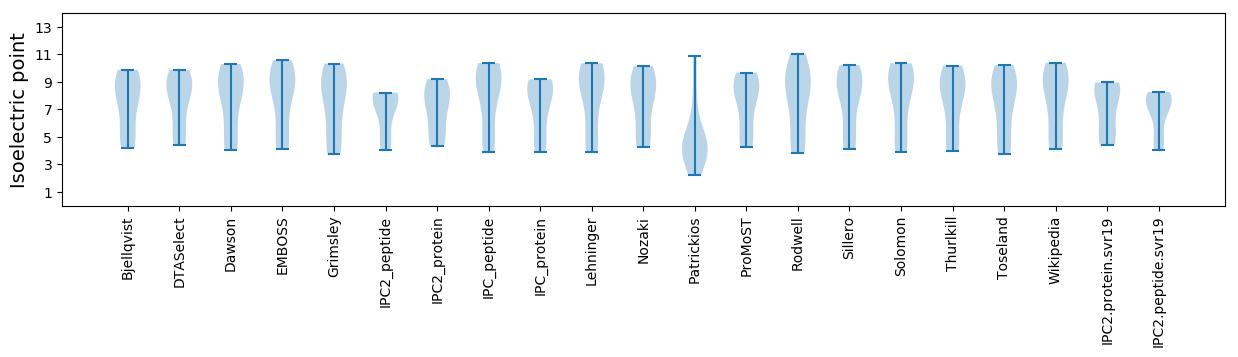
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P03658|G1P_BPIKE Gene 1 protein OS=Salmonella phage IKe OX=10867 GN=I PE=3 SV=1
MM1 pKa = 7.63RR2 pKa = 11.84VLSTVLAAKK11 pKa = 10.51NKK13 pKa = 9.02IALGAATMLVSAGSFAAEE31 pKa = 3.81PNAATNYY38 pKa = 8.56ATEE41 pKa = 4.64AMDD44 pKa = 4.09SLKK47 pKa = 9.34TQAIDD52 pKa = 5.42LISQTWPVVTTVVVAGLVIRR72 pKa = 11.84LFKK75 pKa = 10.7KK76 pKa = 10.83FSSKK80 pKa = 10.85AVV82 pKa = 3.23
MM1 pKa = 7.63RR2 pKa = 11.84VLSTVLAAKK11 pKa = 10.51NKK13 pKa = 9.02IALGAATMLVSAGSFAAEE31 pKa = 3.81PNAATNYY38 pKa = 8.56ATEE41 pKa = 4.64AMDD44 pKa = 4.09SLKK47 pKa = 9.34TQAIDD52 pKa = 5.42LISQTWPVVTTVVVAGLVIRR72 pKa = 11.84LFKK75 pKa = 10.7KK76 pKa = 10.83FSSKK80 pKa = 10.85AVV82 pKa = 3.23
Molecular weight: 8.57 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
2253 |
32 |
437 |
204.8 |
22.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.057 ± 0.591 | 1.198 ± 0.338 |
6.17 ± 0.464 | 3.551 ± 0.815 |
4.749 ± 0.583 | 8.123 ± 1.182 |
1.332 ± 0.33 | 6.303 ± 0.96 |
5.992 ± 0.608 | 8.3 ± 0.858 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.308 ± 0.349 | 5.593 ± 0.82 |
3.24 ± 0.282 | 3.95 ± 0.388 |
3.595 ± 0.412 | 9.498 ± 0.626 |
5.859 ± 0.455 | 8.522 ± 0.856 |
0.976 ± 0.189 | 3.684 ± 0.628 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
