
Bovine faeces associated smacovirus 5
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Drosmacovirus; Drosmacovirus bovas1
Average proteome isoelectric point is 5.35
Get precalculated fractions of proteins
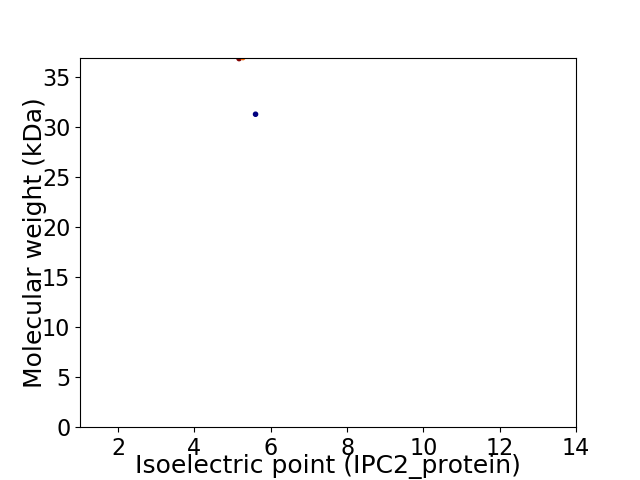
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A168MFL0|A0A168MFL0_9VIRU Putative capsid protein OS=Bovine faeces associated smacovirus 5 OX=1843753 PE=4 SV=1
MM1 pKa = 5.52VTVRR5 pKa = 11.84ISEE8 pKa = 4.33LYY10 pKa = 10.8DD11 pKa = 2.96MKK13 pKa = 11.2TEE15 pKa = 3.72IGKK18 pKa = 9.76IGLLGVHH25 pKa = 6.39TPTGSSIAKK34 pKa = 8.78RR35 pKa = 11.84WHH37 pKa = 5.99GLMINHH43 pKa = 7.04KK44 pKa = 9.25FMRR47 pKa = 11.84IKK49 pKa = 10.67SCDD52 pKa = 3.05IRR54 pKa = 11.84VACAFVLPADD64 pKa = 4.18PLQVGTDD71 pKa = 3.48TTDD74 pKa = 2.97EE75 pKa = 4.2GLIAPQDD82 pKa = 3.26IMNPILYY89 pKa = 9.83RR90 pKa = 11.84AVTNEE95 pKa = 3.03GWNTVINRR103 pKa = 11.84VYY105 pKa = 10.9SSAGMNYY112 pKa = 10.18DD113 pKa = 3.66VNSVLYY119 pKa = 8.18TTDD122 pKa = 3.17AFPSFSEE129 pKa = 4.43GVNDD133 pKa = 3.71LVYY136 pKa = 10.97YY137 pKa = 10.68GLLGQDD143 pKa = 3.91EE144 pKa = 4.58WRR146 pKa = 11.84KK147 pKa = 10.35AMPQAGLSMSGLRR160 pKa = 11.84PLVYY164 pKa = 10.0PLLSSYY170 pKa = 11.51GNTEE174 pKa = 3.06IMEE177 pKa = 4.5PSKK180 pKa = 10.32STSAISYY187 pKa = 9.35PPGQISGQDD196 pKa = 3.5GSSSNPGTNQRR207 pKa = 11.84VPVMRR212 pKa = 11.84GHH214 pKa = 7.18AVPMPRR220 pKa = 11.84LPCTTPGATTVDD232 pKa = 3.94TVPEE236 pKa = 4.14PVFTPQYY243 pKa = 9.64IPKK246 pKa = 9.8SYY248 pKa = 9.75VACIVMPPAKK258 pKa = 9.26MHH260 pKa = 6.34SLFFRR265 pKa = 11.84LAITWYY271 pKa = 10.09IEE273 pKa = 3.9FTDD276 pKa = 4.65LCTALDD282 pKa = 4.23KK283 pKa = 10.14EE284 pKa = 4.74TAGQVFDD291 pKa = 4.89AGDD294 pKa = 3.29YY295 pKa = 8.86TYY297 pKa = 11.2RR298 pKa = 11.84RR299 pKa = 11.84TYY301 pKa = 11.01SFTAGKK307 pKa = 9.23SLKK310 pKa = 10.72DD311 pKa = 3.26IDD313 pKa = 4.05DD314 pKa = 4.01TADD317 pKa = 3.41EE318 pKa = 4.54EE319 pKa = 4.66NSIDD323 pKa = 3.88SLNAPVHH330 pKa = 5.62LVVEE334 pKa = 4.75KK335 pKa = 11.05
MM1 pKa = 5.52VTVRR5 pKa = 11.84ISEE8 pKa = 4.33LYY10 pKa = 10.8DD11 pKa = 2.96MKK13 pKa = 11.2TEE15 pKa = 3.72IGKK18 pKa = 9.76IGLLGVHH25 pKa = 6.39TPTGSSIAKK34 pKa = 8.78RR35 pKa = 11.84WHH37 pKa = 5.99GLMINHH43 pKa = 7.04KK44 pKa = 9.25FMRR47 pKa = 11.84IKK49 pKa = 10.67SCDD52 pKa = 3.05IRR54 pKa = 11.84VACAFVLPADD64 pKa = 4.18PLQVGTDD71 pKa = 3.48TTDD74 pKa = 2.97EE75 pKa = 4.2GLIAPQDD82 pKa = 3.26IMNPILYY89 pKa = 9.83RR90 pKa = 11.84AVTNEE95 pKa = 3.03GWNTVINRR103 pKa = 11.84VYY105 pKa = 10.9SSAGMNYY112 pKa = 10.18DD113 pKa = 3.66VNSVLYY119 pKa = 8.18TTDD122 pKa = 3.17AFPSFSEE129 pKa = 4.43GVNDD133 pKa = 3.71LVYY136 pKa = 10.97YY137 pKa = 10.68GLLGQDD143 pKa = 3.91EE144 pKa = 4.58WRR146 pKa = 11.84KK147 pKa = 10.35AMPQAGLSMSGLRR160 pKa = 11.84PLVYY164 pKa = 10.0PLLSSYY170 pKa = 11.51GNTEE174 pKa = 3.06IMEE177 pKa = 4.5PSKK180 pKa = 10.32STSAISYY187 pKa = 9.35PPGQISGQDD196 pKa = 3.5GSSSNPGTNQRR207 pKa = 11.84VPVMRR212 pKa = 11.84GHH214 pKa = 7.18AVPMPRR220 pKa = 11.84LPCTTPGATTVDD232 pKa = 3.94TVPEE236 pKa = 4.14PVFTPQYY243 pKa = 9.64IPKK246 pKa = 9.8SYY248 pKa = 9.75VACIVMPPAKK258 pKa = 9.26MHH260 pKa = 6.34SLFFRR265 pKa = 11.84LAITWYY271 pKa = 10.09IEE273 pKa = 3.9FTDD276 pKa = 4.65LCTALDD282 pKa = 4.23KK283 pKa = 10.14EE284 pKa = 4.74TAGQVFDD291 pKa = 4.89AGDD294 pKa = 3.29YY295 pKa = 8.86TYY297 pKa = 11.2RR298 pKa = 11.84RR299 pKa = 11.84TYY301 pKa = 11.01SFTAGKK307 pKa = 9.23SLKK310 pKa = 10.72DD311 pKa = 3.26IDD313 pKa = 4.05DD314 pKa = 4.01TADD317 pKa = 3.41EE318 pKa = 4.54EE319 pKa = 4.66NSIDD323 pKa = 3.88SLNAPVHH330 pKa = 5.62LVVEE334 pKa = 4.75KK335 pKa = 11.05
Molecular weight: 36.82 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A168MFL0|A0A168MFL0_9VIRU Putative capsid protein OS=Bovine faeces associated smacovirus 5 OX=1843753 PE=4 SV=1
MM1 pKa = 6.92VRR3 pKa = 11.84SHH5 pKa = 5.89NMEE8 pKa = 3.85RR9 pKa = 11.84ALPDD13 pKa = 3.18EE14 pKa = 5.18HH15 pKa = 6.83GTTQRR20 pKa = 11.84LAYY23 pKa = 9.58QPHH26 pKa = 6.48AKK28 pKa = 9.93EE29 pKa = 3.97GVIGEE34 pKa = 4.59EE35 pKa = 3.92ISPTSGKK42 pKa = 7.2THH44 pKa = 5.71YY45 pKa = 9.18QCKK48 pKa = 7.39WHH50 pKa = 6.82LSRR53 pKa = 11.84GEE55 pKa = 4.78SIDD58 pKa = 3.2GWKK61 pKa = 10.56LLIGPMGHH69 pKa = 6.4VDD71 pKa = 2.83IAVEE75 pKa = 4.13KK76 pKa = 10.56RR77 pKa = 11.84FTGYY81 pKa = 9.37EE82 pKa = 3.92EE83 pKa = 4.76KK84 pKa = 10.54DD85 pKa = 3.3GKK87 pKa = 9.25FVKK90 pKa = 9.89WPEE93 pKa = 3.88SPIAKK98 pKa = 9.66HH99 pKa = 6.38KK100 pKa = 10.87NLALKK105 pKa = 8.23TWEE108 pKa = 4.1VALLDD113 pKa = 4.38SIKK116 pKa = 10.96NQDD119 pKa = 3.53DD120 pKa = 3.43RR121 pKa = 11.84HH122 pKa = 5.28ITVVVDD128 pKa = 3.46KK129 pKa = 11.25QGGNGKK135 pKa = 8.2STFSKK140 pKa = 10.67YY141 pKa = 10.81LEE143 pKa = 4.61ANDD146 pKa = 3.93IADD149 pKa = 3.71VCPVVSDD156 pKa = 4.25EE157 pKa = 4.26YY158 pKa = 11.69NDD160 pKa = 3.51YY161 pKa = 10.74TSYY164 pKa = 11.52CMEE167 pKa = 4.54FPKK170 pKa = 10.35KK171 pKa = 10.46AYY173 pKa = 10.69VFDD176 pKa = 4.44LPRR179 pKa = 11.84ATSIKK184 pKa = 10.03RR185 pKa = 11.84RR186 pKa = 11.84TAMWSGIEE194 pKa = 3.93QIKK197 pKa = 10.96NGLLYY202 pKa = 10.24EE203 pKa = 4.18KK204 pKa = 9.97RR205 pKa = 11.84YY206 pKa = 10.07KK207 pKa = 8.6PRR209 pKa = 11.84KK210 pKa = 7.8MWIEE214 pKa = 3.97PPSILVFTNDD224 pKa = 4.07DD225 pKa = 3.95IPWEE229 pKa = 3.97LLSEE233 pKa = 4.46DD234 pKa = 3.0RR235 pKa = 11.84WDD237 pKa = 3.88AYY239 pKa = 10.69RR240 pKa = 11.84LHH242 pKa = 7.87DD243 pKa = 4.02DD244 pKa = 3.52TLYY247 pKa = 11.27VLSKK251 pKa = 10.8PSDD254 pKa = 3.62DD255 pKa = 4.7YY256 pKa = 11.89INGSNATMDD265 pKa = 3.67EE266 pKa = 4.12DD267 pKa = 4.33SRR269 pKa = 11.84LII271 pKa = 3.85
MM1 pKa = 6.92VRR3 pKa = 11.84SHH5 pKa = 5.89NMEE8 pKa = 3.85RR9 pKa = 11.84ALPDD13 pKa = 3.18EE14 pKa = 5.18HH15 pKa = 6.83GTTQRR20 pKa = 11.84LAYY23 pKa = 9.58QPHH26 pKa = 6.48AKK28 pKa = 9.93EE29 pKa = 3.97GVIGEE34 pKa = 4.59EE35 pKa = 3.92ISPTSGKK42 pKa = 7.2THH44 pKa = 5.71YY45 pKa = 9.18QCKK48 pKa = 7.39WHH50 pKa = 6.82LSRR53 pKa = 11.84GEE55 pKa = 4.78SIDD58 pKa = 3.2GWKK61 pKa = 10.56LLIGPMGHH69 pKa = 6.4VDD71 pKa = 2.83IAVEE75 pKa = 4.13KK76 pKa = 10.56RR77 pKa = 11.84FTGYY81 pKa = 9.37EE82 pKa = 3.92EE83 pKa = 4.76KK84 pKa = 10.54DD85 pKa = 3.3GKK87 pKa = 9.25FVKK90 pKa = 9.89WPEE93 pKa = 3.88SPIAKK98 pKa = 9.66HH99 pKa = 6.38KK100 pKa = 10.87NLALKK105 pKa = 8.23TWEE108 pKa = 4.1VALLDD113 pKa = 4.38SIKK116 pKa = 10.96NQDD119 pKa = 3.53DD120 pKa = 3.43RR121 pKa = 11.84HH122 pKa = 5.28ITVVVDD128 pKa = 3.46KK129 pKa = 11.25QGGNGKK135 pKa = 8.2STFSKK140 pKa = 10.67YY141 pKa = 10.81LEE143 pKa = 4.61ANDD146 pKa = 3.93IADD149 pKa = 3.71VCPVVSDD156 pKa = 4.25EE157 pKa = 4.26YY158 pKa = 11.69NDD160 pKa = 3.51YY161 pKa = 10.74TSYY164 pKa = 11.52CMEE167 pKa = 4.54FPKK170 pKa = 10.35KK171 pKa = 10.46AYY173 pKa = 10.69VFDD176 pKa = 4.44LPRR179 pKa = 11.84ATSIKK184 pKa = 10.03RR185 pKa = 11.84RR186 pKa = 11.84TAMWSGIEE194 pKa = 3.93QIKK197 pKa = 10.96NGLLYY202 pKa = 10.24EE203 pKa = 4.18KK204 pKa = 9.97RR205 pKa = 11.84YY206 pKa = 10.07KK207 pKa = 8.6PRR209 pKa = 11.84KK210 pKa = 7.8MWIEE214 pKa = 3.97PPSILVFTNDD224 pKa = 4.07DD225 pKa = 3.95IPWEE229 pKa = 3.97LLSEE233 pKa = 4.46DD234 pKa = 3.0RR235 pKa = 11.84WDD237 pKa = 3.88AYY239 pKa = 10.69RR240 pKa = 11.84LHH242 pKa = 7.87DD243 pKa = 4.02DD244 pKa = 3.52TLYY247 pKa = 11.27VLSKK251 pKa = 10.8PSDD254 pKa = 3.62DD255 pKa = 4.7YY256 pKa = 11.89INGSNATMDD265 pKa = 3.67EE266 pKa = 4.12DD267 pKa = 4.33SRR269 pKa = 11.84LII271 pKa = 3.85
Molecular weight: 31.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
606 |
271 |
335 |
303.0 |
34.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.941 ± 0.483 | 1.32 ± 0.133 |
7.261 ± 0.765 | 5.611 ± 1.103 |
2.64 ± 0.266 | 6.601 ± 0.434 |
2.475 ± 0.527 | 6.106 ± 0.104 |
5.941 ± 1.588 | 7.426 ± 0.028 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.3 ± 0.447 | 3.795 ± 0.066 |
6.436 ± 0.792 | 2.475 ± 0.163 |
4.62 ± 0.34 | 7.591 ± 0.361 |
6.931 ± 1.1 | 6.766 ± 0.767 |
1.98 ± 0.606 | 4.785 ± 0.007 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
