
Spirosoma oryzae
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Cytophagaceae; Spirosoma
Average proteome isoelectric point is 6.81
Get precalculated fractions of proteins
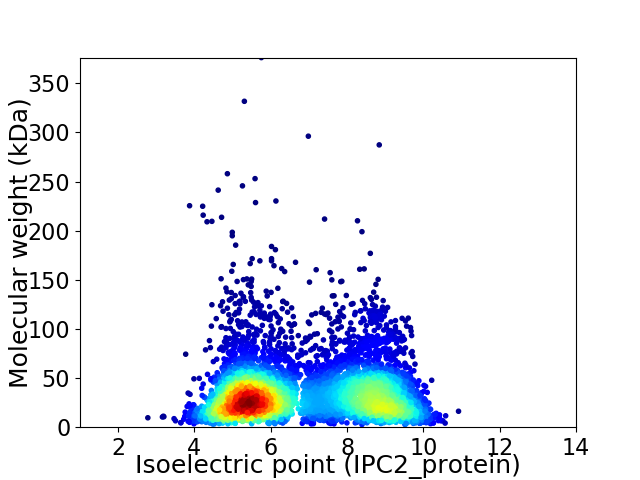
Virtual 2D-PAGE plot for 5655 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
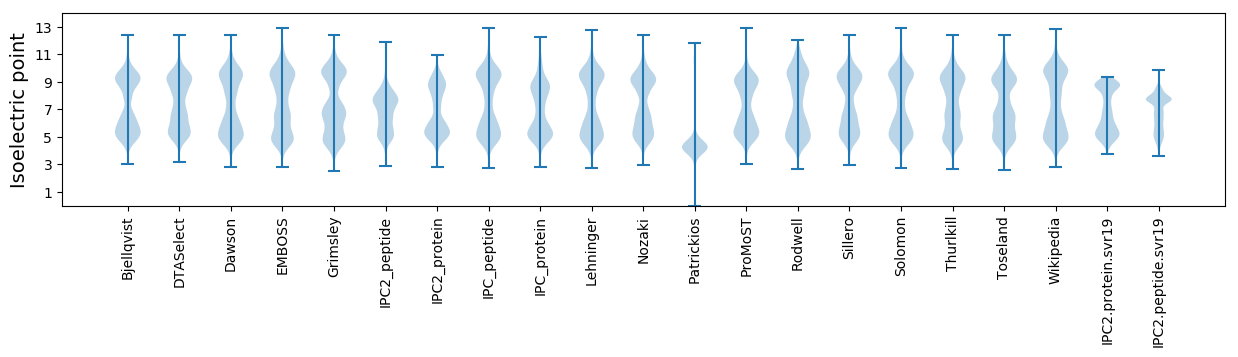
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2T0TIR4|A0A2T0TIR4_9BACT Putative NBD/HSP70 family sugar kinase OS=Spirosoma oryzae OX=1469603 GN=CLV58_102352 PE=3 SV=1
MM1 pKa = 7.66ANYY4 pKa = 9.81KK5 pKa = 10.53NEE7 pKa = 3.92GFDD10 pKa = 3.7PEE12 pKa = 4.97EE13 pKa = 4.89IDD15 pKa = 4.45ALKK18 pKa = 10.58EE19 pKa = 3.63EE20 pKa = 4.63CQAAGKK26 pKa = 10.38SFVYY30 pKa = 10.76VDD32 pKa = 5.36DD33 pKa = 4.77NDD35 pKa = 5.02LDD37 pKa = 3.85VLEE40 pKa = 5.23SGEE43 pKa = 4.32CVHH46 pKa = 6.57IQFPGQYY53 pKa = 9.14QGQEE57 pKa = 4.13VIYY60 pKa = 10.27DD61 pKa = 3.55ALVYY65 pKa = 8.55TLRR68 pKa = 11.84LHH70 pKa = 6.7HH71 pKa = 6.86SSLVYY76 pKa = 10.24EE77 pKa = 4.3MAVEE81 pKa = 4.76EE82 pKa = 4.34VQKK85 pKa = 9.69TYY87 pKa = 10.88PDD89 pKa = 3.47YY90 pKa = 10.61TPPEE94 pKa = 4.05EE95 pKa = 4.93RR96 pKa = 11.84EE97 pKa = 4.06AGYY100 pKa = 10.4KK101 pKa = 9.36IAPDD105 pKa = 3.52RR106 pKa = 11.84EE107 pKa = 4.36EE108 pKa = 4.01EE109 pKa = 4.34AEE111 pKa = 4.12TALTEE116 pKa = 4.32IIGEE120 pKa = 4.16IEE122 pKa = 3.9DD123 pKa = 4.09TEE125 pKa = 4.55TVKK128 pKa = 10.73VQEE131 pKa = 4.93HH132 pKa = 6.44IEE134 pKa = 3.95IDD136 pKa = 3.36TDD138 pKa = 3.23SDD140 pKa = 3.98YY141 pKa = 11.8GVALDD146 pKa = 3.71VCLNTEE152 pKa = 4.74DD153 pKa = 4.4ISDD156 pKa = 3.68EE157 pKa = 4.19VIEE160 pKa = 5.28NFIQQFKK167 pKa = 11.14ANTLALDD174 pKa = 3.47TTLYY178 pKa = 11.01SFTSEE183 pKa = 4.49DD184 pKa = 3.88DD185 pKa = 3.51EE186 pKa = 4.7
MM1 pKa = 7.66ANYY4 pKa = 9.81KK5 pKa = 10.53NEE7 pKa = 3.92GFDD10 pKa = 3.7PEE12 pKa = 4.97EE13 pKa = 4.89IDD15 pKa = 4.45ALKK18 pKa = 10.58EE19 pKa = 3.63EE20 pKa = 4.63CQAAGKK26 pKa = 10.38SFVYY30 pKa = 10.76VDD32 pKa = 5.36DD33 pKa = 4.77NDD35 pKa = 5.02LDD37 pKa = 3.85VLEE40 pKa = 5.23SGEE43 pKa = 4.32CVHH46 pKa = 6.57IQFPGQYY53 pKa = 9.14QGQEE57 pKa = 4.13VIYY60 pKa = 10.27DD61 pKa = 3.55ALVYY65 pKa = 8.55TLRR68 pKa = 11.84LHH70 pKa = 6.7HH71 pKa = 6.86SSLVYY76 pKa = 10.24EE77 pKa = 4.3MAVEE81 pKa = 4.76EE82 pKa = 4.34VQKK85 pKa = 9.69TYY87 pKa = 10.88PDD89 pKa = 3.47YY90 pKa = 10.61TPPEE94 pKa = 4.05EE95 pKa = 4.93RR96 pKa = 11.84EE97 pKa = 4.06AGYY100 pKa = 10.4KK101 pKa = 9.36IAPDD105 pKa = 3.52RR106 pKa = 11.84EE107 pKa = 4.36EE108 pKa = 4.01EE109 pKa = 4.34AEE111 pKa = 4.12TALTEE116 pKa = 4.32IIGEE120 pKa = 4.16IEE122 pKa = 3.9DD123 pKa = 4.09TEE125 pKa = 4.55TVKK128 pKa = 10.73VQEE131 pKa = 4.93HH132 pKa = 6.44IEE134 pKa = 3.95IDD136 pKa = 3.36TDD138 pKa = 3.23SDD140 pKa = 3.98YY141 pKa = 11.8GVALDD146 pKa = 3.71VCLNTEE152 pKa = 4.74DD153 pKa = 4.4ISDD156 pKa = 3.68EE157 pKa = 4.19VIEE160 pKa = 5.28NFIQQFKK167 pKa = 11.14ANTLALDD174 pKa = 3.47TTLYY178 pKa = 11.01SFTSEE183 pKa = 4.49DD184 pKa = 3.88DD185 pKa = 3.51EE186 pKa = 4.7
Molecular weight: 21.14 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2T0RUG1|A0A2T0RUG1_9BACT Zn_protease domain-containing protein OS=Spirosoma oryzae OX=1469603 GN=CLV58_13812 PE=4 SV=1
MM1 pKa = 7.66GEE3 pKa = 4.05LHH5 pKa = 6.03QRR7 pKa = 11.84PSPFRR12 pKa = 11.84IRR14 pKa = 11.84LRR16 pKa = 11.84WVYY19 pKa = 10.2CVVWYY24 pKa = 10.37RR25 pKa = 11.84INCLHH30 pKa = 6.89RR31 pKa = 11.84LSLTDD36 pKa = 3.16PAGSVRR42 pKa = 11.84VGRR45 pKa = 11.84SRR47 pKa = 11.84DD48 pKa = 3.46GQAVVAGPTQGLCPTACSVQKK69 pKa = 10.13STDD72 pKa = 3.39HH73 pKa = 6.75PPEE76 pKa = 4.38LSCLIPCAQWSDD88 pKa = 3.4RR89 pKa = 11.84TVNSQDD95 pKa = 3.73SACLNAVQTKK105 pKa = 7.0CHH107 pKa = 6.72CEE109 pKa = 3.39PGFISRR115 pKa = 11.84QRR117 pKa = 11.84KK118 pKa = 6.04AQRR121 pKa = 11.84IVRR124 pKa = 11.84TVRR127 pKa = 11.84LSAEE131 pKa = 4.12GAAGSIKK138 pKa = 9.71TGRR141 pKa = 11.84PVYY144 pKa = 10.3CNCTTKK150 pKa = 10.73CVGTCLITDD159 pKa = 4.25TASGEE164 pKa = 4.13PIRR167 pKa = 11.84TKK169 pKa = 10.63RR170 pKa = 11.84QLTRR174 pKa = 11.84IDD176 pKa = 3.57RR177 pKa = 11.84AVLGSSSILTVATTIRR193 pKa = 11.84SRR195 pKa = 11.84PRR197 pKa = 11.84SAIVLRR203 pKa = 11.84TTEE206 pKa = 3.93SWVQRR211 pKa = 11.84YY212 pKa = 6.65WVPRR216 pKa = 11.84SRR218 pKa = 4.43
MM1 pKa = 7.66GEE3 pKa = 4.05LHH5 pKa = 6.03QRR7 pKa = 11.84PSPFRR12 pKa = 11.84IRR14 pKa = 11.84LRR16 pKa = 11.84WVYY19 pKa = 10.2CVVWYY24 pKa = 10.37RR25 pKa = 11.84INCLHH30 pKa = 6.89RR31 pKa = 11.84LSLTDD36 pKa = 3.16PAGSVRR42 pKa = 11.84VGRR45 pKa = 11.84SRR47 pKa = 11.84DD48 pKa = 3.46GQAVVAGPTQGLCPTACSVQKK69 pKa = 10.13STDD72 pKa = 3.39HH73 pKa = 6.75PPEE76 pKa = 4.38LSCLIPCAQWSDD88 pKa = 3.4RR89 pKa = 11.84TVNSQDD95 pKa = 3.73SACLNAVQTKK105 pKa = 7.0CHH107 pKa = 6.72CEE109 pKa = 3.39PGFISRR115 pKa = 11.84QRR117 pKa = 11.84KK118 pKa = 6.04AQRR121 pKa = 11.84IVRR124 pKa = 11.84TVRR127 pKa = 11.84LSAEE131 pKa = 4.12GAAGSIKK138 pKa = 9.71TGRR141 pKa = 11.84PVYY144 pKa = 10.3CNCTTKK150 pKa = 10.73CVGTCLITDD159 pKa = 4.25TASGEE164 pKa = 4.13PIRR167 pKa = 11.84TKK169 pKa = 10.63RR170 pKa = 11.84QLTRR174 pKa = 11.84IDD176 pKa = 3.57RR177 pKa = 11.84AVLGSSSILTVATTIRR193 pKa = 11.84SRR195 pKa = 11.84PRR197 pKa = 11.84SAIVLRR203 pKa = 11.84TTEE206 pKa = 3.93SWVQRR211 pKa = 11.84YY212 pKa = 6.65WVPRR216 pKa = 11.84SRR218 pKa = 4.43
Molecular weight: 24.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1917797 |
24 |
3356 |
339.1 |
37.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.703 ± 0.036 | 0.738 ± 0.011 |
5.578 ± 0.025 | 4.698 ± 0.034 |
4.145 ± 0.022 | 7.303 ± 0.04 |
1.78 ± 0.017 | 5.455 ± 0.029 |
4.373 ± 0.038 | 10.163 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.105 ± 0.017 | 4.47 ± 0.032 |
4.811 ± 0.024 | 4.669 ± 0.028 |
5.716 ± 0.032 | 6.209 ± 0.035 |
6.908 ± 0.049 | 7.055 ± 0.029 |
1.295 ± 0.012 | 3.825 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
