
Capybara microvirus Cap1_SP_48
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.23
Get precalculated fractions of proteins
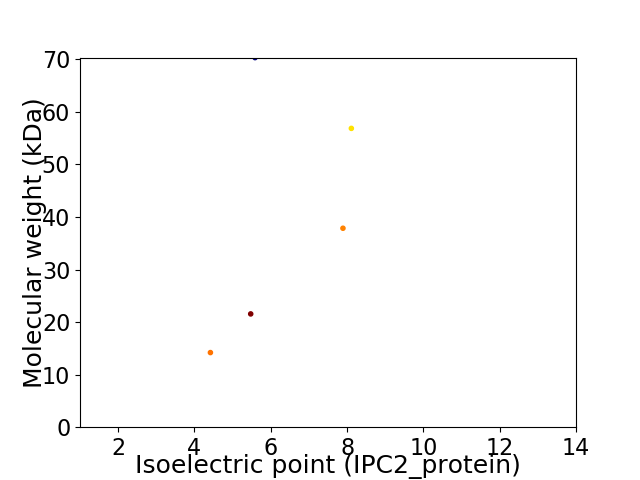
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W4V5|A0A4P8W4V5_9VIRU Minor capsid protein OS=Capybara microvirus Cap1_SP_48 OX=2584779 PE=4 SV=1
MM1 pKa = 7.02NRR3 pKa = 11.84KK4 pKa = 9.25SFIEE8 pKa = 3.94NSVSKK13 pKa = 9.87YY14 pKa = 6.94YY15 pKa = 10.79HH16 pKa = 6.66SIEE19 pKa = 4.41TPPPTNDD26 pKa = 3.39VVCHH30 pKa = 5.36VVDD33 pKa = 3.81VTDD36 pKa = 5.46DD37 pKa = 3.62EE38 pKa = 5.16GNSYY42 pKa = 10.35PVVAIAEE49 pKa = 4.19KK50 pKa = 10.29KK51 pKa = 9.31YY52 pKa = 11.1GSFHH56 pKa = 7.72VSDD59 pKa = 4.82FDD61 pKa = 4.08LTNLINAGIDD71 pKa = 3.46PRR73 pKa = 11.84SVSISQSSAQYY84 pKa = 10.92LDD86 pKa = 5.18DD87 pKa = 4.65IEE89 pKa = 6.16HH90 pKa = 6.92FDD92 pKa = 4.14SLAKK96 pKa = 10.1TILSRR101 pKa = 11.84LDD103 pKa = 3.44NLEE106 pKa = 4.15VSQSPSGEE114 pKa = 4.17TLSIPSPVEE123 pKa = 3.38ATAPSQNN130 pKa = 3.16
MM1 pKa = 7.02NRR3 pKa = 11.84KK4 pKa = 9.25SFIEE8 pKa = 3.94NSVSKK13 pKa = 9.87YY14 pKa = 6.94YY15 pKa = 10.79HH16 pKa = 6.66SIEE19 pKa = 4.41TPPPTNDD26 pKa = 3.39VVCHH30 pKa = 5.36VVDD33 pKa = 3.81VTDD36 pKa = 5.46DD37 pKa = 3.62EE38 pKa = 5.16GNSYY42 pKa = 10.35PVVAIAEE49 pKa = 4.19KK50 pKa = 10.29KK51 pKa = 9.31YY52 pKa = 11.1GSFHH56 pKa = 7.72VSDD59 pKa = 4.82FDD61 pKa = 4.08LTNLINAGIDD71 pKa = 3.46PRR73 pKa = 11.84SVSISQSSAQYY84 pKa = 10.92LDD86 pKa = 5.18DD87 pKa = 4.65IEE89 pKa = 6.16HH90 pKa = 6.92FDD92 pKa = 4.14SLAKK96 pKa = 10.1TILSRR101 pKa = 11.84LDD103 pKa = 3.44NLEE106 pKa = 4.15VSQSPSGEE114 pKa = 4.17TLSIPSPVEE123 pKa = 3.38ATAPSQNN130 pKa = 3.16
Molecular weight: 14.2 kDa
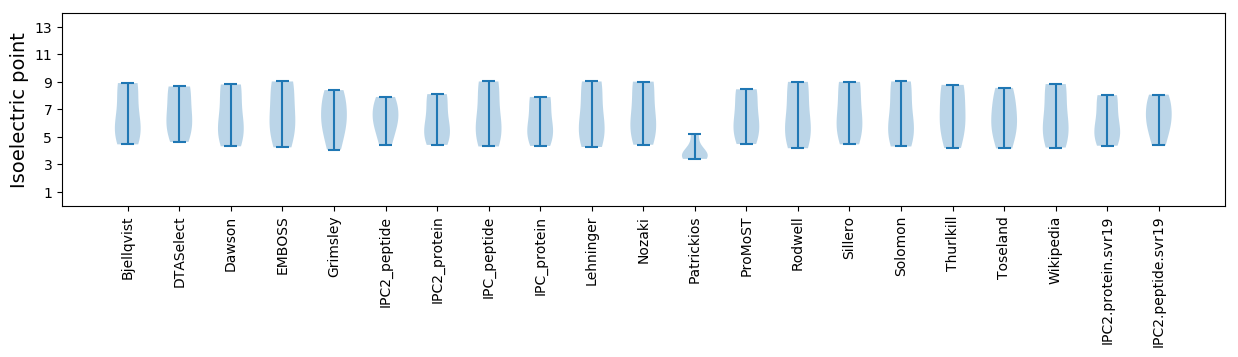
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W7G2|A0A4P8W7G2_9VIRU Peptidase M15 OS=Capybara microvirus Cap1_SP_48 OX=2584779 PE=4 SV=1
MM1 pKa = 7.2RR2 pKa = 11.84TCFSILNRR10 pKa = 11.84SNYY13 pKa = 9.74FDD15 pKa = 3.45IEE17 pKa = 4.09RR18 pKa = 11.84SKK20 pKa = 11.13SHH22 pKa = 7.38FDD24 pKa = 3.18IEE26 pKa = 5.0SNALFLNQLNKK37 pKa = 10.29CNSYY41 pKa = 10.39EE42 pKa = 3.9IRR44 pKa = 11.84NHH46 pKa = 6.59FEE48 pKa = 3.46FEE50 pKa = 4.33EE51 pKa = 4.16IKK53 pKa = 10.93KK54 pKa = 10.79LGGQAFFYY62 pKa = 9.95TLSYY66 pKa = 11.33NDD68 pKa = 2.92AHH70 pKa = 6.84LPHH73 pKa = 6.53FHH75 pKa = 7.61GIPCHH80 pKa = 5.4DD81 pKa = 4.51HH82 pKa = 6.41EE83 pKa = 5.83DD84 pKa = 4.17LRR86 pKa = 11.84ALLNGAFSKK95 pKa = 10.49RR96 pKa = 11.84VKK98 pKa = 10.21RR99 pKa = 11.84YY100 pKa = 8.58CNCSLRR106 pKa = 11.84YY107 pKa = 9.49FITLEE112 pKa = 4.03LGEE115 pKa = 4.68GKK117 pKa = 10.5GSRR120 pKa = 11.84GLFKK124 pKa = 10.71NPHH127 pKa = 4.61YY128 pKa = 10.96HH129 pKa = 6.7CIFYY133 pKa = 10.61LRR135 pKa = 11.84PKK137 pKa = 10.23EE138 pKa = 3.83GEE140 pKa = 4.04IYY142 pKa = 11.09NKK144 pKa = 9.68IDD146 pKa = 3.75PLTFRR151 pKa = 11.84SFIRR155 pKa = 11.84EE156 pKa = 3.61YY157 pKa = 9.46WQGVDD162 pKa = 3.84IDD164 pKa = 4.66FSHH167 pKa = 7.44PSARR171 pKa = 11.84AKK173 pKa = 10.52DD174 pKa = 3.86FRR176 pKa = 11.84FGICQEE182 pKa = 3.66GKK184 pKa = 10.09YY185 pKa = 10.19FGEE188 pKa = 4.27ILTSAACRR196 pKa = 11.84YY197 pKa = 7.96CCKK200 pKa = 10.03YY201 pKa = 9.0TIKK204 pKa = 10.78SNAVIQHH211 pKa = 5.52YY212 pKa = 9.2LSCVSEE218 pKa = 4.08IQRR221 pKa = 11.84DD222 pKa = 3.69FDD224 pKa = 3.83NSLGGIKK231 pKa = 10.49YY232 pKa = 9.81SVSPEE237 pKa = 4.07EE238 pKa = 4.13YY239 pKa = 10.43NSFIDD244 pKa = 3.85DD245 pKa = 3.39QVKK248 pKa = 10.06IFKK251 pKa = 9.95NRR253 pKa = 11.84YY254 pKa = 6.32FVKK257 pKa = 10.17VRR259 pKa = 11.84CSQGLGSYY267 pKa = 10.42GLSQVNDD274 pKa = 3.85DD275 pKa = 4.4KK276 pKa = 11.94VVVPYY281 pKa = 10.82KK282 pKa = 10.18STFKK286 pKa = 10.65SYY288 pKa = 10.17PIPLYY293 pKa = 10.28YY294 pKa = 9.98YY295 pKa = 9.66RR296 pKa = 11.84KK297 pKa = 10.19LYY299 pKa = 10.0TNHH302 pKa = 6.69CKK304 pKa = 9.99NALGNVHH311 pKa = 6.62YY312 pKa = 9.53YY313 pKa = 10.66HH314 pKa = 6.81NSKK317 pKa = 10.43GLEE320 pKa = 3.76RR321 pKa = 11.84LRR323 pKa = 11.84NTLHH327 pKa = 6.92LRR329 pKa = 11.84IDD331 pKa = 3.71KK332 pKa = 10.81AKK334 pKa = 10.66SDD336 pKa = 3.73ALKK339 pKa = 10.37LVSMLSDD346 pKa = 3.2KK347 pKa = 11.14DD348 pKa = 3.89FNLSFTRR355 pKa = 11.84WLEE358 pKa = 3.99SNGIILSSSILDD370 pKa = 4.12LGLCAQRR377 pKa = 11.84YY378 pKa = 9.08AEE380 pKa = 4.11FSVVYY385 pKa = 10.15KK386 pKa = 10.59NRR388 pKa = 11.84YY389 pKa = 6.9YY390 pKa = 10.89TDD392 pKa = 4.25LNCPIDD398 pKa = 3.6IDD400 pKa = 4.21LDD402 pKa = 3.75HH403 pKa = 7.35EE404 pKa = 4.5FFIIPVEE411 pKa = 4.26TEE413 pKa = 3.74NALHH417 pKa = 6.86RR418 pKa = 11.84LAVQKK423 pKa = 10.63GRR425 pKa = 11.84YY426 pKa = 7.05EE427 pKa = 3.87VHH429 pKa = 6.74SYY431 pKa = 11.44ANHH434 pKa = 7.18PYY436 pKa = 9.39FSPYY440 pKa = 9.84YY441 pKa = 10.47VQFCLLDD448 pKa = 3.62LLKK451 pKa = 10.91EE452 pKa = 4.42FFRR455 pKa = 11.84QTISTTMYY463 pKa = 9.91RR464 pKa = 11.84KK465 pKa = 9.67KK466 pKa = 10.55CAIEE470 pKa = 3.63EE471 pKa = 4.18TRR473 pKa = 11.84RR474 pKa = 11.84VHH476 pKa = 6.82LSRR479 pKa = 11.84RR480 pKa = 11.84FPSS483 pKa = 3.19
MM1 pKa = 7.2RR2 pKa = 11.84TCFSILNRR10 pKa = 11.84SNYY13 pKa = 9.74FDD15 pKa = 3.45IEE17 pKa = 4.09RR18 pKa = 11.84SKK20 pKa = 11.13SHH22 pKa = 7.38FDD24 pKa = 3.18IEE26 pKa = 5.0SNALFLNQLNKK37 pKa = 10.29CNSYY41 pKa = 10.39EE42 pKa = 3.9IRR44 pKa = 11.84NHH46 pKa = 6.59FEE48 pKa = 3.46FEE50 pKa = 4.33EE51 pKa = 4.16IKK53 pKa = 10.93KK54 pKa = 10.79LGGQAFFYY62 pKa = 9.95TLSYY66 pKa = 11.33NDD68 pKa = 2.92AHH70 pKa = 6.84LPHH73 pKa = 6.53FHH75 pKa = 7.61GIPCHH80 pKa = 5.4DD81 pKa = 4.51HH82 pKa = 6.41EE83 pKa = 5.83DD84 pKa = 4.17LRR86 pKa = 11.84ALLNGAFSKK95 pKa = 10.49RR96 pKa = 11.84VKK98 pKa = 10.21RR99 pKa = 11.84YY100 pKa = 8.58CNCSLRR106 pKa = 11.84YY107 pKa = 9.49FITLEE112 pKa = 4.03LGEE115 pKa = 4.68GKK117 pKa = 10.5GSRR120 pKa = 11.84GLFKK124 pKa = 10.71NPHH127 pKa = 4.61YY128 pKa = 10.96HH129 pKa = 6.7CIFYY133 pKa = 10.61LRR135 pKa = 11.84PKK137 pKa = 10.23EE138 pKa = 3.83GEE140 pKa = 4.04IYY142 pKa = 11.09NKK144 pKa = 9.68IDD146 pKa = 3.75PLTFRR151 pKa = 11.84SFIRR155 pKa = 11.84EE156 pKa = 3.61YY157 pKa = 9.46WQGVDD162 pKa = 3.84IDD164 pKa = 4.66FSHH167 pKa = 7.44PSARR171 pKa = 11.84AKK173 pKa = 10.52DD174 pKa = 3.86FRR176 pKa = 11.84FGICQEE182 pKa = 3.66GKK184 pKa = 10.09YY185 pKa = 10.19FGEE188 pKa = 4.27ILTSAACRR196 pKa = 11.84YY197 pKa = 7.96CCKK200 pKa = 10.03YY201 pKa = 9.0TIKK204 pKa = 10.78SNAVIQHH211 pKa = 5.52YY212 pKa = 9.2LSCVSEE218 pKa = 4.08IQRR221 pKa = 11.84DD222 pKa = 3.69FDD224 pKa = 3.83NSLGGIKK231 pKa = 10.49YY232 pKa = 9.81SVSPEE237 pKa = 4.07EE238 pKa = 4.13YY239 pKa = 10.43NSFIDD244 pKa = 3.85DD245 pKa = 3.39QVKK248 pKa = 10.06IFKK251 pKa = 9.95NRR253 pKa = 11.84YY254 pKa = 6.32FVKK257 pKa = 10.17VRR259 pKa = 11.84CSQGLGSYY267 pKa = 10.42GLSQVNDD274 pKa = 3.85DD275 pKa = 4.4KK276 pKa = 11.94VVVPYY281 pKa = 10.82KK282 pKa = 10.18STFKK286 pKa = 10.65SYY288 pKa = 10.17PIPLYY293 pKa = 10.28YY294 pKa = 9.98YY295 pKa = 9.66RR296 pKa = 11.84KK297 pKa = 10.19LYY299 pKa = 10.0TNHH302 pKa = 6.69CKK304 pKa = 9.99NALGNVHH311 pKa = 6.62YY312 pKa = 9.53YY313 pKa = 10.66HH314 pKa = 6.81NSKK317 pKa = 10.43GLEE320 pKa = 3.76RR321 pKa = 11.84LRR323 pKa = 11.84NTLHH327 pKa = 6.92LRR329 pKa = 11.84IDD331 pKa = 3.71KK332 pKa = 10.81AKK334 pKa = 10.66SDD336 pKa = 3.73ALKK339 pKa = 10.37LVSMLSDD346 pKa = 3.2KK347 pKa = 11.14DD348 pKa = 3.89FNLSFTRR355 pKa = 11.84WLEE358 pKa = 3.99SNGIILSSSILDD370 pKa = 4.12LGLCAQRR377 pKa = 11.84YY378 pKa = 9.08AEE380 pKa = 4.11FSVVYY385 pKa = 10.15KK386 pKa = 10.59NRR388 pKa = 11.84YY389 pKa = 6.9YY390 pKa = 10.89TDD392 pKa = 4.25LNCPIDD398 pKa = 3.6IDD400 pKa = 4.21LDD402 pKa = 3.75HH403 pKa = 7.35EE404 pKa = 4.5FFIIPVEE411 pKa = 4.26TEE413 pKa = 3.74NALHH417 pKa = 6.86RR418 pKa = 11.84LAVQKK423 pKa = 10.63GRR425 pKa = 11.84YY426 pKa = 7.05EE427 pKa = 3.87VHH429 pKa = 6.74SYY431 pKa = 11.44ANHH434 pKa = 7.18PYY436 pKa = 9.39FSPYY440 pKa = 9.84YY441 pKa = 10.47VQFCLLDD448 pKa = 3.62LLKK451 pKa = 10.91EE452 pKa = 4.42FFRR455 pKa = 11.84QTISTTMYY463 pKa = 9.91RR464 pKa = 11.84KK465 pKa = 9.67KK466 pKa = 10.55CAIEE470 pKa = 3.63EE471 pKa = 4.18TRR473 pKa = 11.84RR474 pKa = 11.84VHH476 pKa = 6.82LSRR479 pKa = 11.84RR480 pKa = 11.84FPSS483 pKa = 3.19
Molecular weight: 56.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1773 |
130 |
624 |
354.6 |
40.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.979 ± 1.378 | 1.805 ± 0.53 |
5.697 ± 0.561 | 5.133 ± 0.566 |
5.979 ± 0.711 | 6.091 ± 0.791 |
2.538 ± 0.683 | 5.64 ± 0.546 |
5.753 ± 0.444 | 8.404 ± 0.474 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.143 ± 0.715 | 6.43 ± 0.842 |
3.553 ± 0.478 | 3.892 ± 0.932 |
4.625 ± 0.681 | 9.081 ± 0.779 |
5.302 ± 0.746 | 6.035 ± 0.617 |
0.79 ± 0.283 | 5.133 ± 1.196 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
