
Truepera radiovictrix (strain DSM 17093 / CIP 108686 / LMG 22925 / RQ-24)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Deinococcus-Thermus; Deinococci; Trueperales; Trueperaceae; Truepera; Truepera radiovictrix
Average proteome isoelectric point is 6.71
Get precalculated fractions of proteins
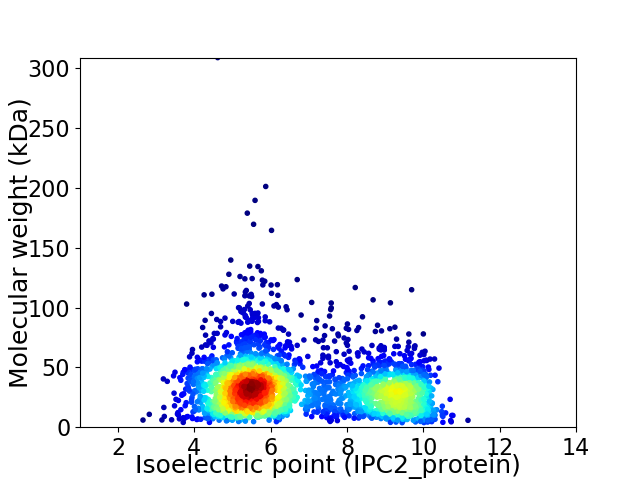
Virtual 2D-PAGE plot for 2924 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D7CQH3|D7CQH3_TRURR Oxidoreductase domain protein OS=Truepera radiovictrix (strain DSM 17093 / CIP 108686 / LMG 22925 / RQ-24) OX=649638 GN=Trad_1841 PE=4 SV=1
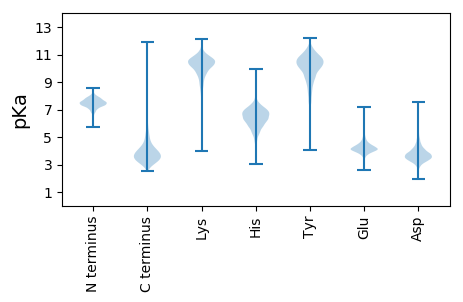
MM1 pKa = 7.36KK2 pKa = 9.08RR3 pKa = 11.84TLATATLLAGTLLGAASAQDD23 pKa = 3.74APSGAITVTAWDD35 pKa = 3.46IAAEE39 pKa = 4.0ALEE42 pKa = 3.99ALVPGFNEE50 pKa = 3.92QYY52 pKa = 10.33PDD54 pKa = 3.49VQVTVEE60 pKa = 4.04NLGNQQVFDD69 pKa = 4.52RR70 pKa = 11.84GLAGCAAGGADD81 pKa = 4.55LPDD84 pKa = 3.63VYY86 pKa = 11.15AVEE89 pKa = 4.21NNEE92 pKa = 5.75AEE94 pKa = 4.59VFWNRR99 pKa = 11.84FPDD102 pKa = 3.85CFLDD106 pKa = 4.08LTALGADD113 pKa = 4.21EE114 pKa = 5.64LRR116 pKa = 11.84DD117 pKa = 3.98DD118 pKa = 4.93FPDD121 pKa = 4.55FKK123 pKa = 10.04WTEE126 pKa = 3.91LTVGEE131 pKa = 4.33RR132 pKa = 11.84VYY134 pKa = 10.89AIPWDD139 pKa = 3.78SGPVVTFYY147 pKa = 10.99RR148 pKa = 11.84RR149 pKa = 11.84DD150 pKa = 3.39LYY152 pKa = 11.14EE153 pKa = 3.95EE154 pKa = 4.86AGINPDD160 pKa = 5.29EE161 pKa = 4.93IEE163 pKa = 3.96TWDD166 pKa = 3.89DD167 pKa = 4.0FIEE170 pKa = 4.71AGQALDD176 pKa = 3.59EE177 pKa = 4.32ATGAKK182 pKa = 9.09MLWFDD187 pKa = 4.85FSDD190 pKa = 3.88DD191 pKa = 4.04GVWRR195 pKa = 11.84QLANQNGCFYY205 pKa = 10.57FDD207 pKa = 3.82PEE209 pKa = 4.38GTEE212 pKa = 3.87VTVNQPGCVDD222 pKa = 2.97AMEE225 pKa = 4.43TMKK228 pKa = 10.97ALYY231 pKa = 10.03DD232 pKa = 3.69AGIVAIVDD240 pKa = 3.31WNGAIQEE247 pKa = 4.23IRR249 pKa = 11.84NGTLAGAVFGAWYY262 pKa = 10.47VGTIQSNAPDD272 pKa = 3.51QEE274 pKa = 4.84GNWGVYY280 pKa = 8.3EE281 pKa = 4.39MPASEE286 pKa = 4.31PGGVRR291 pKa = 11.84AANLGGSALAIPASSQNPEE310 pKa = 3.26AAMAFIRR317 pKa = 11.84YY318 pKa = 8.76ALATTEE324 pKa = 4.55GQVSMLRR331 pKa = 11.84NQGLVPSYY339 pKa = 10.89LPALEE344 pKa = 4.7DD345 pKa = 4.18PYY347 pKa = 11.12VQEE350 pKa = 4.49PVPYY354 pKa = 10.01WGDD357 pKa = 3.33QPIWQDD363 pKa = 2.79ILATLGEE370 pKa = 4.31IPQARR375 pKa = 11.84GTQYY379 pKa = 10.35FQEE382 pKa = 4.11ARR384 pKa = 11.84QIVTNTLSQYY394 pKa = 8.32LTQNAYY400 pKa = 10.13EE401 pKa = 4.42DD402 pKa = 3.88AQAAMDD408 pKa = 4.99DD409 pKa = 3.68AAQQISAAVGLPIAGQQ425 pKa = 3.22
MM1 pKa = 7.36KK2 pKa = 9.08RR3 pKa = 11.84TLATATLLAGTLLGAASAQDD23 pKa = 3.74APSGAITVTAWDD35 pKa = 3.46IAAEE39 pKa = 4.0ALEE42 pKa = 3.99ALVPGFNEE50 pKa = 3.92QYY52 pKa = 10.33PDD54 pKa = 3.49VQVTVEE60 pKa = 4.04NLGNQQVFDD69 pKa = 4.52RR70 pKa = 11.84GLAGCAAGGADD81 pKa = 4.55LPDD84 pKa = 3.63VYY86 pKa = 11.15AVEE89 pKa = 4.21NNEE92 pKa = 5.75AEE94 pKa = 4.59VFWNRR99 pKa = 11.84FPDD102 pKa = 3.85CFLDD106 pKa = 4.08LTALGADD113 pKa = 4.21EE114 pKa = 5.64LRR116 pKa = 11.84DD117 pKa = 3.98DD118 pKa = 4.93FPDD121 pKa = 4.55FKK123 pKa = 10.04WTEE126 pKa = 3.91LTVGEE131 pKa = 4.33RR132 pKa = 11.84VYY134 pKa = 10.89AIPWDD139 pKa = 3.78SGPVVTFYY147 pKa = 10.99RR148 pKa = 11.84RR149 pKa = 11.84DD150 pKa = 3.39LYY152 pKa = 11.14EE153 pKa = 3.95EE154 pKa = 4.86AGINPDD160 pKa = 5.29EE161 pKa = 4.93IEE163 pKa = 3.96TWDD166 pKa = 3.89DD167 pKa = 4.0FIEE170 pKa = 4.71AGQALDD176 pKa = 3.59EE177 pKa = 4.32ATGAKK182 pKa = 9.09MLWFDD187 pKa = 4.85FSDD190 pKa = 3.88DD191 pKa = 4.04GVWRR195 pKa = 11.84QLANQNGCFYY205 pKa = 10.57FDD207 pKa = 3.82PEE209 pKa = 4.38GTEE212 pKa = 3.87VTVNQPGCVDD222 pKa = 2.97AMEE225 pKa = 4.43TMKK228 pKa = 10.97ALYY231 pKa = 10.03DD232 pKa = 3.69AGIVAIVDD240 pKa = 3.31WNGAIQEE247 pKa = 4.23IRR249 pKa = 11.84NGTLAGAVFGAWYY262 pKa = 10.47VGTIQSNAPDD272 pKa = 3.51QEE274 pKa = 4.84GNWGVYY280 pKa = 8.3EE281 pKa = 4.39MPASEE286 pKa = 4.31PGGVRR291 pKa = 11.84AANLGGSALAIPASSQNPEE310 pKa = 3.26AAMAFIRR317 pKa = 11.84YY318 pKa = 8.76ALATTEE324 pKa = 4.55GQVSMLRR331 pKa = 11.84NQGLVPSYY339 pKa = 10.89LPALEE344 pKa = 4.7DD345 pKa = 4.18PYY347 pKa = 11.12VQEE350 pKa = 4.49PVPYY354 pKa = 10.01WGDD357 pKa = 3.33QPIWQDD363 pKa = 2.79ILATLGEE370 pKa = 4.31IPQARR375 pKa = 11.84GTQYY379 pKa = 10.35FQEE382 pKa = 4.11ARR384 pKa = 11.84QIVTNTLSQYY394 pKa = 8.32LTQNAYY400 pKa = 10.13EE401 pKa = 4.42DD402 pKa = 3.88AQAAMDD408 pKa = 4.99DD409 pKa = 3.68AAQQISAAVGLPIAGQQ425 pKa = 3.22
Molecular weight: 45.94 kDa
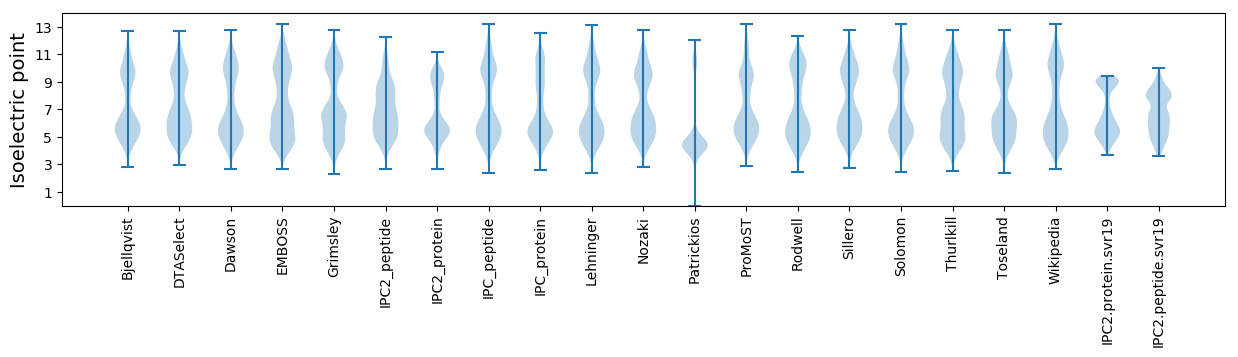
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D7CTU7|D7CTU7_TRURR Uncharacterized protein OS=Truepera radiovictrix (strain DSM 17093 / CIP 108686 / LMG 22925 / RQ-24) OX=649638 GN=Trad_2538 PE=4 SV=1
MM1 pKa = 7.56KK2 pKa = 10.5LNRR5 pKa = 11.84FLRR8 pKa = 11.84TALKK12 pKa = 10.76LFVLTRR18 pKa = 11.84GFRR21 pKa = 11.84AARR24 pKa = 11.84RR25 pKa = 11.84GVGRR29 pKa = 11.84YY30 pKa = 8.31GGRR33 pKa = 11.84RR34 pKa = 11.84VRR36 pKa = 11.84GGVVQQLMRR45 pKa = 11.84RR46 pKa = 11.84LLRR49 pKa = 4.36
MM1 pKa = 7.56KK2 pKa = 10.5LNRR5 pKa = 11.84FLRR8 pKa = 11.84TALKK12 pKa = 10.76LFVLTRR18 pKa = 11.84GFRR21 pKa = 11.84AARR24 pKa = 11.84RR25 pKa = 11.84GVGRR29 pKa = 11.84YY30 pKa = 8.31GGRR33 pKa = 11.84RR34 pKa = 11.84VRR36 pKa = 11.84GGVVQQLMRR45 pKa = 11.84RR46 pKa = 11.84LLRR49 pKa = 4.36
Molecular weight: 5.76 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
943825 |
35 |
3080 |
322.8 |
34.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.344 ± 0.062 | 0.652 ± 0.012 |
4.592 ± 0.039 | 6.623 ± 0.043 |
3.696 ± 0.028 | 8.829 ± 0.041 |
2.02 ± 0.024 | 3.182 ± 0.041 |
2.151 ± 0.035 | 12.421 ± 0.08 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.609 ± 0.018 | 2.027 ± 0.024 |
5.859 ± 0.035 | 2.904 ± 0.022 |
8.02 ± 0.047 | 4.678 ± 0.028 |
5.402 ± 0.031 | 8.107 ± 0.039 |
1.355 ± 0.018 | 2.528 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
